A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes (original) (raw)
SNPs are common in the human genome and often provide correlative evidence for the involvement of specific genes in human disease1. SNPs that affect the function of crucial components of the T-cell antigen receptor (TCR) signaling pathways could have profound effects on the function of the immune system and thus the development of autoimmune diseases2. Protein tyrosine phosphatases3 (PTPs) are particularly good candidates for carrying disease-related SNPs because they are involved in preventing spontaneous T-cell activation and they restrict the response to antigen by dephosphorylating and inactivating TCR-associated kinases and their substrates4,5. PTPs are also needed to revert activated T blasts to a resting phenotype6.
The lymphoid-specific phosphatase (LYP) encoded by PTPN22 is a 110-kDa PTP consisting of an N-terminal phosphatase domain and a long noncatalytic C terminus with several proline-rich motifs4,7. LYP is expressed in lymphocytes7, where it physically associates through its most N-terminal proline-rich motif (called P1) with the SH3 domain of the Csk kinase8, an important suppressor of the Src family kinases Lck9 and Fyn, which mediate TCR signaling. LYP is among the most powerful inhibitors of T-cell activation4,10, a task accomplished by dephosphorylation of TCR-associated kinases, like Lck4, Fyn10 and ZAP-70 (ref. 10). Dephosphorylation of ZAP-70 may be connected to the negative regulatory function of the c-Cbl proto-oncogene, as LYP forms a complex with c-Cbl7.
We found that PTPN22 contains a SNP at nucleotide 1858 in codon 620 (Supplementary Fig. 1 online), which encodes an arginine (CGG) in most donors and in all published human and mouse LYP sequences, but encodes a tryptophan (TGG) in at least one allele of PTPN22 in ∼16.2% of genotyped healthy, non-Hispanic white individuals from the North American population (Table 1). Only 4 of 395 healthy individuals were homozygous with respect to the PTPN22 1858T allele. Because of the proposed role of LYP as a gatekeeper in T-cell activation, we determined whether the SNP was correlated with autoimmune disease.
Table 1 Frequency of PTPN22 genotypes in individuals with T1D in two independent samples
Insulin-dependent type 1 diabetes mellitus (T1D) affects 0.5% of the human population and ∼1.4 million people in the United States. The disease is believed to arise as the consequence of an autoimmune destruction of insulin-producing β-cells by cytotoxic CD8+ T cells with help from CD4+ T cells. First, we carried out a genetic association study using a sample of 294 North American individuals with T1D and 395 healthy subjects from the same ethnic background. We genotyped these individuals and found that the PTPN22 allele 1858T was more frequent in individuals with T1D than in healthy individuals: 30.6% of individuals with T1D were heterozygous with respect to 1858T, compared with only 21.3% of the healthy controls, and 3.7% of individuals with T1D were homozygous with respect to 1858T, compared with only 1.0% of the healthy controls (χ2 = 14.84 with 2 degrees of freedom, P < 0.001; odds ratio for the 1858T carriers versus noncarriers = 1.83, 95% confidence interval = 1.284–2.596). As the cohorts were matched for age and race, these results suggest that the PTPN22 allele 1858T predisposes an individual to developing T1D.
This result was supported by genotyping a second, completely independent and ethnically separate cohort of 388 individuals from Sardinia, Italy. All affected individuals and controls came from northern Sardinia, a region characterized by an ethnically homogenous population. In this study, more than twice as many individuals with T1D as controls were heterozygous with respect to 1858T (8.6% versus 4.2%, respectively) and the only individual who was homozygous with respect to 1858T in the cohort had the disease (χ2 = 3.93 with 1 degree of freedom, P < 0.05; odds ratio for the 1858T carriers versus noncarriers = 2.31, 95% confidence interval = 0.932–5.819). This confirms that the PTPN22 allele 1858T confers predisposition to T1D, even in a different background in which the allele frequency is lower. As LYP is expressed in lymphocytes and the disease is caused by lymphocytes, this correlation probably relates to altered lymphocyte function. Chromosomal region 1p13, where the PTPN22 gene resides, is also weakly linked to systemic lupus erythematosus11 and rheumatoid arthritis12, suggesting that it might be worth addressing a possible role for LYP in systemic autoimmune diseases.
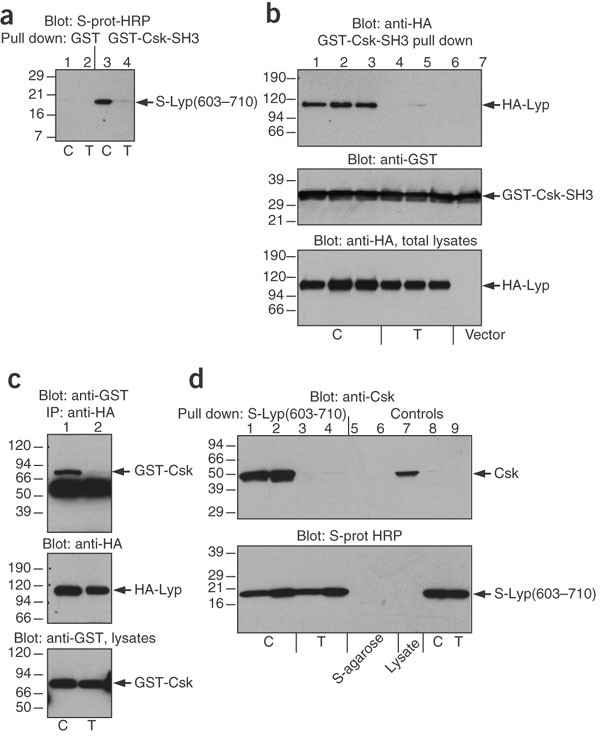
To gain some insight into the possible molecular basis of the disease correlation, we examined the location and role of the amino acid encoded by codon 620. This residue resides in the P1 proline-rich motif involved in binding to the SH3 domain of Csk (Supplementary Fig. 1 online). The three-dimensional structure of the complex13 shows that the side chain of Arg620 fits into a somewhat acidic depression within the ligand binding cleft of the SH3 domain, where it interacts with a tryptophan (Trp47) of Csk (Supplementary Fig. 1 online). Replacing Arg620 with a tryptophan residue is expected to disrupt this interaction. The side chain of tryptophan is also bulkier and would not fit into the pocket. Indeed, when we expressed amino acids 603–710 of LYP as an S-tagged protein in Escherichia coli, only the Arg620 construct was precipitated by the SH3 domain of Csk (Fig. 1a). Similarly, full-length LYP with Arg620 (allele 1858C) expressed in COS cells was readily precipitated by the Csk SH3 domain, whereas LYP with Trp620 (allele 1858T) was not (Fig. 1b). Full-length LYP with Arg620 and Csk also coimmunoprecipitated (Fig. 1c), and endogenous Csk was readily precipitated from T cells by S-tagged LYP peptide 603–710 with Arg620 but not Trp620 (Fig. 1d). Together, these results indicate that only LYP with Arg620 (allele 1858C) forms a complex with the Csk kinase, whereas LYP with Trp620 (allele 1858T) does not.
Figure 1: LYP with Trp620 (allele 1858T) does not bind Csk.
(a) Binding of an S-tagged LYP polypeptide corresponding to amino acid residues 603–710 with Arg620 (allele 1858C) or Trp620 (allele 1858T) to GST (lanes 1 and 2) or GST-Csk-SH3 (lanes 3 and 4). The bound material was detected with S-protein-horseradish peroxidase (HRP). Both forms of the peptide were equally expressed in E. coli BL21. (b) Immunoblotting with antibody to hemagglutinin (HA; top panel) of material precipitated with GST-Csk-SH3 from COS cells transfected with three different amounts of plasmid expressing LYP with either Arg620 or Trp620. Blotting with antibody to GST of the same filter (middle panel) and blotting with antibody to hemagglutinin (HA) of lysates from the same transfectants (bottom panel) show equal expression of the two LYP variants. (c) Immunoblotting with antibody to GST (top panel) of material immunoprecipitated with LYP from COS cells transfected with GST-Csk and the two LYP proteins. Blotting with antibody to hemagglutinin (HA; middle panel) and with antibody to GST (bottom panel) of lysates from the same transfectants show equal expression. (d) Immunoblotting with antibody to Csk (upper panel) of endogenous Csk precipitated with the S-tagged LYP(603–710) protein with either Arg620 or Trp620 from Jurkat T cells. Lane 6 is material bound to S-agarose alone, lane 7 is total lysates of Jurkat cells and lanes 8 and 9 are the S-tagged protein alone. S-protein-HRP blot of the same samples (lower panel). Expression of recombinant proteins, protein binding, transfections, immunoprecipitation and immunoblotting were done as described14,15.
Our findings suggest that a SNP in lymphoid tyrosine phosphatase is part of the complex genetic background of a common autoimmune disease, T1D. The SNP directly disrupts the formation of the LYP-Csk complex, the physiological relevance of which is well documented4,10. The mouse ortholog of LYP (called PEP) did not inhibit T-cell activation in an antigen-specific T-cell hybridoma when Csk association was disrupted by mutation of the P1 motif10. We found that mouse4 as well as human (data not shown) LYP inhibits activation of human Jurkat T cells in a synergistic manner with Csk4. Thus, individuals heterozygous with respect to the PTPN22 allele 1858T may have reduced amounts of LYP-Csk complexes, and homozygous individuals no such complexes. Therefore, hyper-reactive T cells are more prone to mount a destructive immune response against autoantigens after viral infection or other immune stresses.
Note: Supplementary information is available on the Nature Genetics website.