Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A (original) (raw)
Charcot-Marie-Tooth disease (CMT) comprises a frequently occurring, genetically heterogeneous group of peripheral neuropathies, although the clinical picture is rather uniform1. Following electrophysiological criteria, CMT falls into two main forms: the demyelinating CMT type 1 with decreased nerve conduction velocities and the axonal form, CMT type 2. In contrast to the well-known molecular genetic defects causing the CMT1 phenotype, the genes associated with CMT2A, CMT2B, CMT2D and CMT2E have only recently been identified. A mutation in KIF1B was previously reported to be associated with CMT2A2, but no further mutations in KIF1B have been identified, although several families with different ethnic origins with linkage to the CMT2A locus on 1p36.2 have been reported3,4,5,6.
We included seven families with CMT2A with the classic clinical phenotype and different ethnic backgrounds in the present study: families DUK662 (ref. 3), DUK1241 and DUK170 (ref. 5), CMT156 (ref. 6), RU45 (identified by I.V.M, E.L.D. and O.E), CMT166 (identified by N.B-T and E.N.), and J693 (ref. 4). We found lod scores between 1.9 and 5.88, indicating linkage to the CMT2A locus (Table 1). Direct sequencing of the entire KIF1B gene, encompassing the two splice variants α and β, identified no mutation in the coding exons of the affected individuals in all families but identified several intronic and synonymous single-nucleotide polymorphisms distributed over the entire gene (Supplementary Table 1 online). We found no mutation in KIF1B cDNA of families CMT156 and CMT166.
Table 1 Clinical, ethnic and genetic characterization of families with CMT2A
This led us to investigate additional genes in the 9.6-cM chromosomal region associated with CMT2A. We excluded mutations in 14 candidate genes with known expression in the nervous system (UBE4B, PEX14, TARDBP, PMSCL2, FRAP1, KIAA1337, FBXO2, FBX30, FBXO6, CLCN6, NPPA, NPPB, TNFRSF8 and VPS13D). But all seven families with CMT2A had missense mutations in the gene encoding mitofusin 2 (MFN2), which is located 1.65 Mb centromeric of KIF1B (Fig. 1 and Table 1). MFN2 is ubiquitously expressed7,8,9, and we identified mRNA transcripts in spinal cord and peripheral nerve (Supplementary Fig. 1 online). MFN2 is localized to the outer mitochondrial membrane and regulates the mitochondrial network architecture by fusion of mitochondria7,8,9,10,11. All the mutations that we detected in MFN2 cosegregated with the disease phenotype in the respective families. None of the resulting amino acid changes were detected in 250 healthy control samples (500 chromosomes) of European descent or in 70 additional Japanese controls (140 chromosomes). Moreover, in 36 families with axonal CMT, each too small for linkage analysis, we identified seven individuals (19%) with additional MFN2 mutations. Most of these individuals had childhood-onset CMT, including one who was diagnosed at the age of one year (Supplementary Table 2 online).
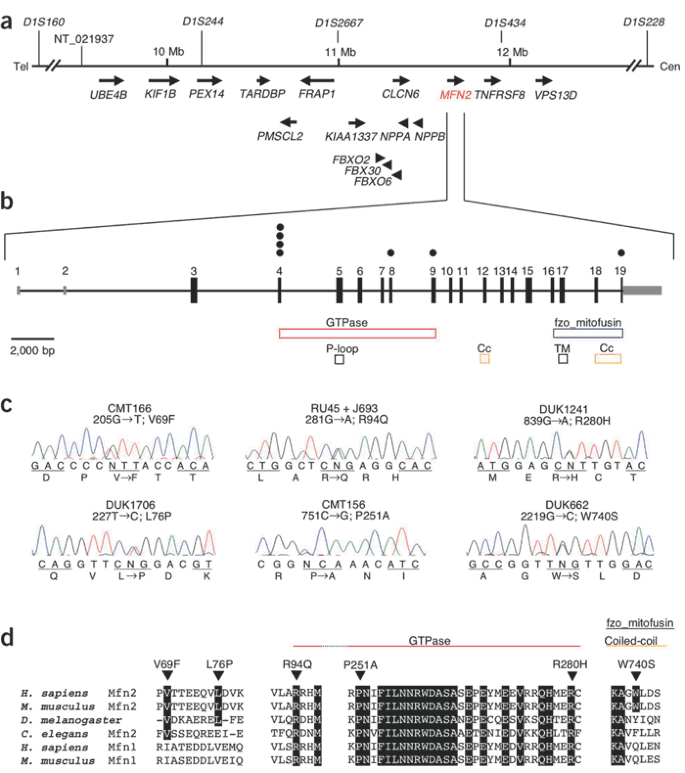
Figure 1: Overview of chromosomal region 1p36.2, the genomic organization of MFN2, the detected mutations and their conservation in different species.
(a) Transcript map of the region on chromosome 1p36.2 associated with CMT2A. The physical map of contig NT_021937 contains KIF1B, typical STR markers and the screened genes including MFN2. The markers D1S160 and D1S434 define the CMT2A-linked locus. The exons of the entire KIF1B gene as well as cDNA transcripts were amplified and directly sequenced following standard procedures. Primer sequences are available on request. Tel, telomeric; Cen, centromeric. (b) Genomic structure of MFN2 with six detected missense mutations in seven families (filled circles). Functional domains (colored bars), translated mRNAs (black bars) and untranslated mRNAs (gray bars) are indicated. TM, transmembrane domain; Cc, coiled-coil domain. (c) Direct sequencing of the amplified coding exons identified six different missense mutations in seven families with CMT2A. Table 1 presents details of these mutations and the affected families. We amplified DNA of family members in standard PCR reactions (primer sequences available on request). and sequenced it using the ABI system (Applied Biosystems). (d) The affected amino acids are highly conserved between different species and within the homologous protein mitofusin 1 as indicated by triangles and black background. Most mutations fell into the GTPase domain, but one was found in the fzo_mitofusin domain. The D. melanogaster homolog is called mitochondrial assembly regulatory factor.
The amino acids affected by the MFN2 mutations in the seven families with CMT2A are highly conserved in different species, including Caenorhabditis elegans and Drosophila melanogaster (Fig. 1d; the D. melanogaster homolog is called fuzzy onions, fzo). Six of seven identified mutations are within or immediately upstream of the GTPase domain of MFN2 (Fig. 1b,d). Four mutations were clustered in exon 4. Both the Russian family RU45 and the Japanese kindred J693 carried the mutation 281G→A, resulting in the amino acid substitution R94Q. Arg94 marks the predicted beginning of the GTPase domain and is conserved in the GTPase domain of mitofusin 1, the only human homolog with an fzo_mitofusin domain (Fig. 1d). An intact GTPase domain is essential for the function of mitofusins7,10,11. In family DUK662, the 2219G→C transversion led to a substitution of the aromatic tryptophan to the small polar serine (W740S). This exchange was predicted to extend the coiled-coil domain at the end of the fzo_mitofusin domain (Supplementary Fig. 2 online).
Mitochondria undergo a dynamically regulated balance between fusion and fission reactions and have a tubular and branched membrane network12. An efficient mitochondrial network is required for fundamental cell functions, such as equilibrating mitochondrial gene products to overcome acquired somatic mutations of mitochondrial DNA13 and establishing a uniform membrane potential at the mitochondrial double membrane for even energy supply throughout the cell14. In addition, studies have implicated mitochondrial dynamics in the regulation of apoptosis. Mitofusin 2 might be involved in this process, as it colocalizes with the proapoptotic protein Bax15.
Homozygous Mfn2 knockout mice die in midgestation owing to placental defects11. Although heterozygotes were reported to have a normal phenotype, mouse embryonic fibroblast cultures from _Mfn2_-deficient mice had markedly lower mitochondrial mobility11. Mobility and transport of mitochondria are key elements to the functional health of the extended neuronal axons, particularly in peripheral nerves. This could be a clue to a possible mechanism of action in CMT2A. Further study may be needed to see if Mfn2 +/− mice develop an abnormal phenotype with age.
Introducing a virally transported mitofusin 2 construct in a _Mfn2_-deficient mouse cell line rescued the normal phenotype by correcting the fusion-fission imbalance11. This raises the possibility that CMT2A may be amenable to some similar type of intervention in the future.
We conclude that mutations in MFN2 are the primary cause underlying CMT2A. The present study demonstrates a new mechanism for axonal neuropathies and should provide insight into the pathophysiology of neuropathic disease, both hereditary and acquired.
Accession numbers. Entrez Protein: Homo sapiens mitofusin 2, AAH17061; Mus musculus mitofusin 2, AAM88577, D. melanogaster mitochondrial assembly regulatory factor, AAM00196; C. elegans mitofusin 2, NP_495161; H. sapiens mitofusin 1, AAH40557; M. musculus mitofusin 1, NP-077162.
GenBank: H. sapiens chromosome 1 genomic contig, NT_021937; UBE4B, NM_006048; PEX14, NM_004565; TARDBP, NM_007375; PMSCL2, NM_002685; FRAP1, NM_004958; KIAA1337, XM_052561; FBXO2, NM_012168; FBX30, NM_033182; FBXO6, NM_018438; CLCN6, NM_001286; NPPA, NM_006172; NPPB, NM_002521; TNFRSF8, NM_001243; VPS13D, XM_044546; KIF1B, NM_015074; MFN2, NM_014874; MFN1, NM_033540.
Note: Supplementary information is available on the Nature Genetics website.