Modularity and evolutionary constraint on proteins (original) (raw)
- Brief Communication
- Published: 06 March 2005
Nature Genetics volume 37, pages 351–352 (2005)Cite this article
- 1455 Accesses
- 172 Citations
- Metrics details
Abstract
Modularity, which has been found in the functional and physical protein interaction networks of many organisms, has been postulated to affect both the mode and tempo of evolution. Here I show that in the yeast Saccharomyces cerevisiae, protein interaction hubs situated in single modules are highly constrained, whereas those connecting different modules are more plastic. This pattern of change could reflect a tendency for evolutionary innovations to occur by altering the proteins and interactions between rather than within modules, in a manner somewhat similar to the evolution of new proteins through the shuffling of conserved protein domains.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout
Additional access options:
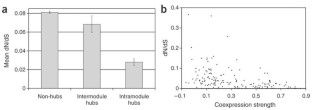
Figure 1: Intramodule hubs are more evolutionarily constrained than intermodule hubs.
Similar content being viewed by others
References
- Needham, J. Biol. Rev. 8, 180–233 (1933).
Article Google Scholar - Gerhart, J. & Kirschner, M. Cells, Embryos, and Evolution (Blackwell Science, Malden, Massachusetts, 1997).
Google Scholar - Hartwell, L.H., Hopfield, J.J., Leibler, S. & Murray, A.W. Nature 402, C47–C52 (1999).
Article CAS Google Scholar - Schlosser, G. Theory Biosci. 121, 1–80 (2002).
Article Google Scholar - Schlosser, G. & Wagner, G.P. (eds.) Modularity in Development and Evolution (University of Chicago Press, Chicago, Illinois, 2004).
Google Scholar - Fisher, R.A. The Genetical Theory of Natural Selection (Dover, New York, New York, 1930).
Book Google Scholar - Waxman, D. & Peck, J.R. Science 279, 1210–1213 (1998).
Article CAS Google Scholar - von Mering, C. et al. Nature 417, 399–403 (2002).
Article CAS Google Scholar - Han, J.D. et al. Nature 430, 88–93 (2004).
Article CAS Google Scholar - Hirsh, A.E., Fraser, H.B. & Wall, D.P. Mol. Biol. Evol. 22, 174–177 (2005).
Article CAS Google Scholar - Fraser, H.B., Hirsh, A.E., Steinmetz, L.M., Scharfe, C. & Feldman, M.W. Science 296, 750–752 (2002).
Article CAS Google Scholar - Jordan, I.K., Marino-Ramirez, L., Wolf, Y.I. & Koonin, E.V. Mol. Biol. Evol. 21, 2058–2070 (2004).
Article CAS Google Scholar - Krylov, D.M., Wolf, Y.I., Rogozin, I.B. & Koonin, E.V. Genome Res. 13, 2229–2235 (2003).
Article CAS Google Scholar - Doolittle, R.F. Annu. Rev. Biochem. 64, 287–314 (1995).
Article CAS Google Scholar - Fraser, H.B., Hirsh, A.E., Wall, D.P. & Eisen, M.B. Proc. Natl. Acad. Sci. USA 101, 9033–9038 (2004).
Article CAS Google Scholar
Acknowledgements
I thank J. Plotkin and A. Hirsh for suggestions. This work was supported by a National Science Foundation predoctoral fellowship.
Author information
Authors and Affiliations
- Department of Molecular and Cell Biology, University of California, Berkeley, 94720, California, USA
Hunter B Fraser
Corresponding author
Correspondence toHunter B Fraser.
Ethics declarations
Competing interests
The author declares no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Fraser, H. Modularity and evolutionary constraint on proteins.Nat Genet 37, 351–352 (2005). https://doi.org/10.1038/ng1530
- Received: 04 October 2004
- Accepted: 03 February 2005
- Published: 06 March 2005
- Issue date: 01 April 2005
- DOI: https://doi.org/10.1038/ng1530