GeneProf: analysis of high-throughput sequencing experiments (original) (raw)
- Correspondence
- Published: 28 December 2011
Nature Methods volume 9, pages 7–8 (2012)Cite this article
- 2086 Accesses
- 74 Citations
- 7 Altmetric
- Metrics details
To the Editor:
The huge volume and complexity of data produced by high-throughput sequencing make it difficult for researchers in many laboratories to fully harness the potential of these data for the study of biological processes and human disease. Data processing rather than generation is now often the bottleneck for biological experiments1, and the efficient use of high-throughput sequencing data submitted to public databases such as the Sequence Read Archive remains a challenging goal for many. Workflow-based software2,3 offers an attractive approach for dealing with complex data because it allows the visual organization of software components into ordered 'workflows' (Supplementary Note). This enables complicated analyses without any need to write custom computer scripts. However, workflow engines focus on the mechanics of the computational processes involved; the primary goal is to achieve computation rather than a particular biological result. Therefore, setting up a workflow can be a daunting task for many life scientists, especially those lacking experience in the visual programming paradigm. Existing tools are hence not sufficient to make high-throughput sequencing data fully accessible to the entire research community.
This is a preview of subscription content, access via your institution
Relevant articles
Open Access articles citing this article.
RNAdetector: a free user-friendly stand-alone and cloud-based system for RNA-Seq data analysis
- Alessandro La Ferlita
- , Salvatore Alaimo
- … Alfredo Pulvirenti
BMC Bioinformatics Open Access 03 June 2021
DolphinNext: a distributed data processing platform for high throughput genomics
- Onur Yukselen
- , Osman Turkyilmaz
- … Alper Kucukural
BMC Genomics Open Access 19 April 2020
Two factor-based reprogramming of rodent and human fibroblasts into Schwann cells
- Pietro Giuseppe Mazzara
- , Luca Massimino
- … Vania Broccoli
Nature Communications Open Access 07 February 2017
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Additional access options:
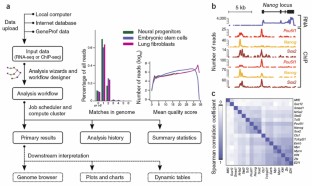
Figure 1: Overview of a GeneProf experiment and sample outputs.
References
- Mardis, E.R. Genome Med. 2, 84 (2010).
Article Google Scholar - Goecks, J., Nekrutenko, A. & Taylor, J. Genome Biol. 11, R86 (2010).
Article Google Scholar - Hull, D. et al. Nucleic Acids Res. 34, W729–W732 (2006).
Article CAS Google Scholar - Ioannidis, J.P. et al. Nat. Genet. 41, 149–155 (2009).
Article CAS Google Scholar - Piwowar, H.A., Day, R.S. & Fridsma, D.B. PLoS One 2, e308 (2007).
Article Google Scholar - Guttman, M. et al. Nat. Biotechnol. 28, 503–510 (2010).
Article CAS Google Scholar - Chen, X. et al. Cell 133, 1106–1117 (2008).
Article CAS Google Scholar - Marson, A. et al. Cell 134, 521–533 (2008).
Article CAS Google Scholar
Acknowledgements
We thank C. Nerlov, I. Chambers and S. Skylaki for proofreading the manuscript and providing helpful suggestions. F.H. was funded by a Medical Research Council studentship. Additional funding was provided by the European Commission Seventh Framework Programme 'EuroSyStem'.
Author information
Authors and Affiliations
- Institute for Stem Cell Research, Centre for Regenerative Medicine, School of Biological Sciences, University of Edinburgh, Edinburgh, UK
Florian Halbritter, Harsh J Vaidya & Simon R Tomlinson
Authors
- Florian Halbritter
- Harsh J Vaidya
- Simon R Tomlinson
Corresponding author
Correspondence toSimon R Tomlinson.
Ethics declarations
Competing interests
Edinburgh Research and Innovation (University of Edinburgh) is currently investigating the commercial potential of GeneProf.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9, Supplementary Note, Supplementary Discussion, Supplementary Methods, Supplementary Data (PDF 5119 kb)
Rights and permissions
About this article
Cite this article
Halbritter, F., Vaidya, H. & Tomlinson, S. GeneProf: analysis of high-throughput sequencing experiments.Nat Methods 9, 7–8 (2012). https://doi.org/10.1038/nmeth.1809
- Published: 28 December 2011
- Issue Date: January 2012
- DOI: https://doi.org/10.1038/nmeth.1809