Microhomology-based choice of Cas9 nuclease target sites (original) (raw)
- Correspondence
- Published: 27 June 2014
Nature Methods volume 11, pages 705–706 (2014)Cite this article
- 17k Accesses
- 355 Citations
- 13 Altmetric
- Metrics details
Subjects
To the Editor:
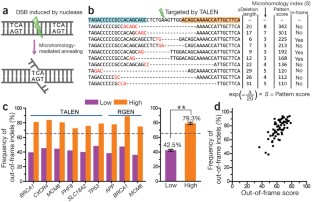
First we examined the mutations induced by ten TALENs and ten RGENs in human cells via deep sequencing (Supplementary Table 1 and Supplementary Methods). We focused our analysis on deletions because they are much more prevalent than insertions (98.7% vs. 1.3%, respectively, for TALENs; 75.1% vs. 24.9% for RGENs) and because microhomology is irrelevant for insertions. In aggregate, microhomologies of 2–8 bases were found in 44.3% and 52.7% of all deletions induced by TALENs and RGENs, respectively (Supplementary Fig. 1 and Supplementary Table 2). Thus, 43.7% (0.987 × 0.443) and 39.6% (0.751 × 0.527) of all the mutations induced by TALENs and RGENs, respectively, were associated with microhomology. At a given nuclease target site, the effect of these microhomology-associated deletions can be predicted. In the extreme cases, (i) all deletions cause frameshifts in a protein-coding gene or (ii) no deletions cause frameshifts. In contrast, one-third of microhomology-independent deletions result in in-frame mutations. Assuming that ∼60% of indels are microhomology independent on average, the fraction of in-frame mutations at a given site can range from 20% (60%/3 + 0%) to 60% (60%/3 + 40%), a threefold difference between the two extreme cases. Because most eukaryotic cells are diploid rather than haploid, the fraction of null cells carrying two out-of-frame mutations can range from 16% (0.40 × 0.40) to 64% (0.80 × 0.80), depending on the choice of target site.
This is a preview of subscription content, access via your institution
Relevant articles
Open Access articles citing this article.
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Additional access options:
Figure 1: Prediction of nuclease-induced deletion patterns associated with microhomology.
References
- Cho, S.W. et al. Nat. Biotechnol. 31, 230–232 (2013).
Article CAS Google Scholar - Kim, H. & Kim, J.-S. Nat. Rev. Genet. 15, 321–334 (2014).
Article CAS Google Scholar - Morton, J. et al. Proc. Natl. Acad. Sci. USA 103, 16370–16375 (2006).
Article CAS Google Scholar - Qi, Y. et al. Genome Res. 23, 547–554 (2013).
Article CAS Google Scholar - Sung, Y.H. et al. Nat. Biotechnol. 31, 23–24 (2013).
Article CAS Google Scholar - Sung, Y.H. et al. Genome Res. 24, 125–131 (2014).
Article CAS Google Scholar - Bae, S. et al. Bioinformatics 30, 1473–1475 (2014).
Article CAS Google Scholar
Acknowledgements
We thank J. Park for the help with website construction. This work was supported by the Plant Molecular Breeding Center of Next-Generation BioGreen 21 Program (PJ009081), the National Research Foundation of Korea (2013065262) and TJ Park Science Fellowship to S.B.
Author information
Authors and Affiliations
- Center for Genome Engineering, Institute for Basic Science, Seoul, South Korea
Sangsu Bae, Jiyeon Kweon, Heon Seok Kim & Jin-Soo Kim - Department of Chemistry, Seoul National University, Seoul, South Korea
Sangsu Bae, Jiyeon Kweon, Heon Seok Kim & Jin-Soo Kim
Authors
- Sangsu Bae
- Jiyeon Kweon
- Heon Seok Kim
- Jin-Soo Kim
Corresponding author
Correspondence toJin-Soo Kim.
Ethics declarations
Competing interests
S.B. and J.-S.K. have filed a patent application based on this work.
Supplementary information
Rights and permissions
About this article
Cite this article
Bae, S., Kweon, J., Kim, H. et al. Microhomology-based choice of Cas9 nuclease target sites.Nat Methods 11, 705–706 (2014). https://doi.org/10.1038/nmeth.3015
- Published: 27 June 2014
- Issue Date: July 2014
- DOI: https://doi.org/10.1038/nmeth.3015