Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques (original) (raw)
Main
The rapid expansion of the COVID-19 pandemic has made the development of a SARS-CoV-2 vaccine a global health priority. Ad26 vectors11 that encode viral antigens have been shown to induce robust humoral and cellular immune responses to various pathogens in both non-human primates and humans. In this study, we developed a series of Ad26 vectors that encode different variants of the SARS-CoV-2 spike (S) protein, and evaluated their immunogenicity and protective efficacy against SARS-CoV-2 challenge in rhesus macaques.
Generation and immunogenicity of Ad26 vectors
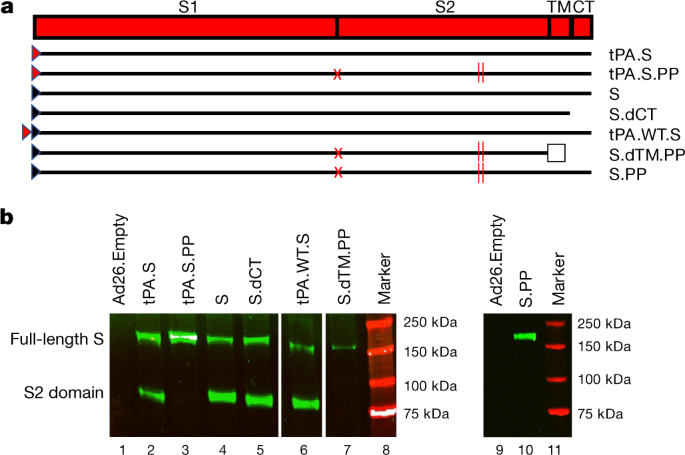
We produced seven Ad26 vectors that expressed SARS-CoV-2 S variants that reflected different leader sequences, antigen forms and stabilization mutations: (i) tissue plasminogen activator (tPA) leader sequence with full-length S (tPA.S)12; (ii) tPA leader sequence with full-length S and mutation of the furin cleavage site and two proline-stabilizing mutations (tPA.S.PP)13,14,15; (iii) wild-type leader sequence with native full-length S (S); (iv) wild-type leader sequence with S and deletion of the cytoplasmic tail (S.dCT)16; (v) tandem tPA and wild-type leader sequences with full-length S (tPA.WT.S)12; (vi) wild-type leader sequence with S with deletion of the transmembrane region and cytoplasmic tail reflecting the soluble ectodomain, with mutation of the furin cleavage site, proline-stabilizing mutations, and a foldon trimerization domain (S.dTM.PP)15; and (vii) wild-type leader sequence with full-length S and mutation of the furin cleavage site and proline-stabilizing mutations (S.PP) (Fig. 1a). Western blot analyses confirmed expression of S in cell lysates from all vectors (Fig. 1b).
Fig. 1: Construction of Ad26 vectors.
a, Seven Ad26 vectors were produced that expressed SARS-CoV-2 S protein variants: (i) tPA leader sequence with full-length S (tPA.S)12; (ii) tPA leader sequence with full-length S with mutation of the furin cleavage site and two proline stabilizing mutations (tPA.S.PP)13,14,15; (iii) wild-type leader sequence with native full-length S (S); (iv) wild-type leader sequence with S with deletion of the cytoplasmic tail (S.dCT)16; (v) tandem tPA and wild-type leader sequences with full-length S as a strategy to enhance expression (tPA.WT.S)12; (vi) wild-type leader sequence with S with deletion of the transmembrane region and cytoplasmic tail, reflecting the soluble ectodomain, with mutation of the furin cleavage site, proline stabilizing mutations and a foldon trimerization domain (S.dTM.PP)15; and (vii) wild-type leader sequence with full-length S with mutation of the furin cleavage site and proline stabilizing mutations (S.PP). The red triangle depicts tPA leader sequence; black triangle depicts wild-type leader sequence; red X depicts furin cleavage site mutation; red vertical lines depict proline mutations; and open square depicts foldon trimerization domain. CT, cytoplasmic domain; S1 and S2, the first and second domain of the S protein; TM, transmembrane region. b, Western blot analyses for expression from Ad26 vaccine vectors encoding tPA.S (lane 2), tPA.S.PP (lane 3), S (lane 4), S.dCT (lane 5), tPA.WT.S (lane 6), S.dTM.PP (lane 7) or S.PP (lane 9) under non-reducing conditions in human MRC-5 cell lysates using a human monoclonal antibody (CR3046). This experiment was repeated three times. For gel source data, see Supplementary Fig. 1.
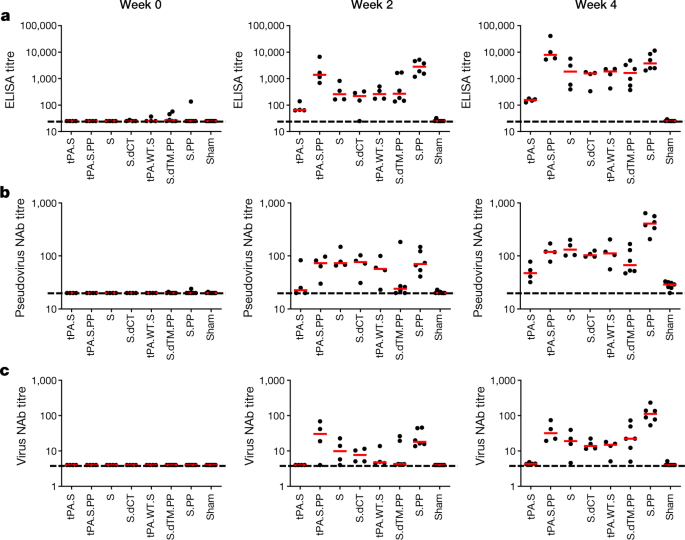
We immunized 52 adult rhesus macaques, aged 6–12 years old, with Ad26 vectors expressing tPA.S (n = 4), tPA.S.PP (N = 4), S (n = 4), S.dCT (n = 4), tPA.WT.S (n = 4), S.dTM.PP (n = 6) or S.PP (n = 6), and sham controls (n = 20). Macaques received a single immunization of 1 × 1011 viral particles of Ad26 vectors by the intramuscular route without adjuvant at week 0. We observed receptor-binding-domain (RBD)-specific binding antibodies by enzyme-linked immunosorbent assay (ELISA) in 31 out of 32 vaccinated macaques by week 2, and in all vaccinated macaques by week 4 (Fig. 2a). Neutralizing antibody responses were assessed using both a pseudovirus neutralization assay9,10,16 (Fig. 2b) and a live virus neutralization assay9,10,17,18 (Fig. 2c). Titres of neutralizing antibodies as measured by both assays were observed in most vaccinated macaques at week 2 and generally increased by week 4. The Ad26-S.PP vaccine elicited the highest titres of pseudovirus neutralizing antibodies (median 408, range 208–643) and live virus neutralizing antibodies (median 113, range 53–233) at week 4 (P < 0.05, two-sided Mann–Whitney tests). Titres of pseudovirus neutralizing antibodies correlated with both ELISA titres and live virus neutralizing antibody titres (P < 0.0001, R = 0.8314 and P < 0.0001, R = 0.8427, respectively, two-sided Spearman rank-correlation tests) (Extended Data Fig. 1). Median titres of neutralizing antibodies in the macaques vaccinated with Ad26-S.PP were fourfold higher than median titres of neutralizing antibodies in previously reported cohorts of 9 convalescent macaques9 and 27 convalescent humans after recovery from SARS-CoV-2 infection10 (P < 0.0001, two-sided Mann–Whitney test) (Extended Data Fig. 2a). The Ad26-S.PP vaccine also induced detectable S-specific IgG and IgA responses in bronchoalveolar lavage (BAL) samples (Extended Data Fig. 2b).
Fig. 2: Humoral immune responses in vaccinated rhesus macaques.
a–c, Humoral immune responses were assessed at weeks 0, 2 and 4 by a RBD-specific binding antibody ELISA (a), pseudovirus neutralization assays (b), and live virus neutralization assays (c). Red bars reflect median responses. Dotted lines reflect assay limit of quantification. NAb, neutralizing antibody.
We further characterized S-specific and RBD-specific antibody responses in the vaccinated macaques by systems serology19. A variety of Fc effector functions, including antibody-dependent neutrophil phagocytosis (ADNP), antibody-dependent monocyte cellular phagocytosis (ADCP), antibody-dependent complement deposition (ADCD) and antibody-dependent natural killer cell activation (ADNKA), as well as several Ig subclasses and FcR binding were observed (Extended Data Fig. 3). The highest antibody-binding responses were observed with the Ad26-tPA.S.PP and Ad26-S.PP vaccines, and the highest effector function responses were seen with the Ad26-S.PP vaccine. A principal component analysis showed substantial overlap of the groups, although Ad26-S.PP was the most divergent group (Extended Data Fig. 3).
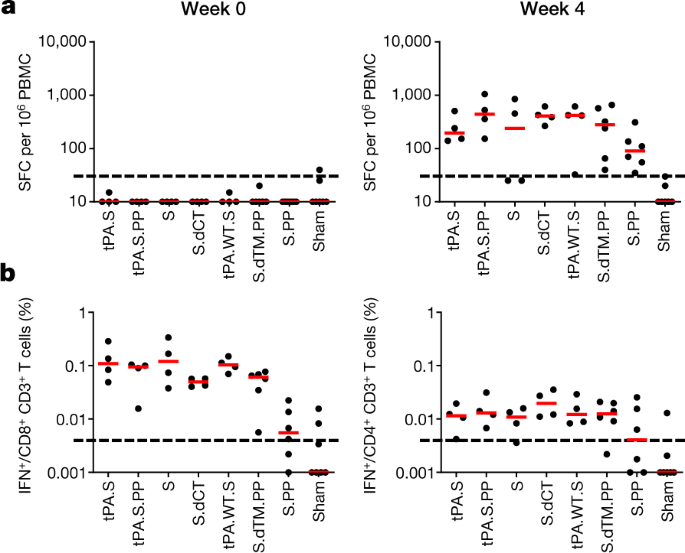
Cellular immune responses were induced in 30 out of 32 vaccinated macaques at week 4 by IFNγ enzyme-linked immune absorbent spot (ELISPOT) assays using pooled S peptides (Fig. 3a), and multiparameter intracellular cytokine staining assays were used to assess IFNγ+CD4+ and CD8+ T cell responses (Fig. 3b). Responses were comparable across the vaccine groups. Analysis of a cohort of 10 similarly immunized macaques demonstrated that a single immunization of 1 × 1011 viral particles of Ad26-S.PP elicited consistent IFNγ ELISPOT responses but minimal or no IL-4 ELISPOT responses (Extended Data Fig. 4), which suggests induction of type 1 T helper (TH1)-biased responses.
Fig. 3: Cellular immune responses in vaccinated rhesus macaques.
a, b, Cellular immune responses were assessed at week 4 after immunization by IFNγ ELISPOT assays (a) and IFNγ+CD4+ and IFNγ+CD8+ T cell intracellular cytokine staining assays (b) in response to pooled S peptides. Red bars reflect median responses. Dotted lines reflect assay limit of quantification.
Protective efficacy of Ad26 vaccine candidates
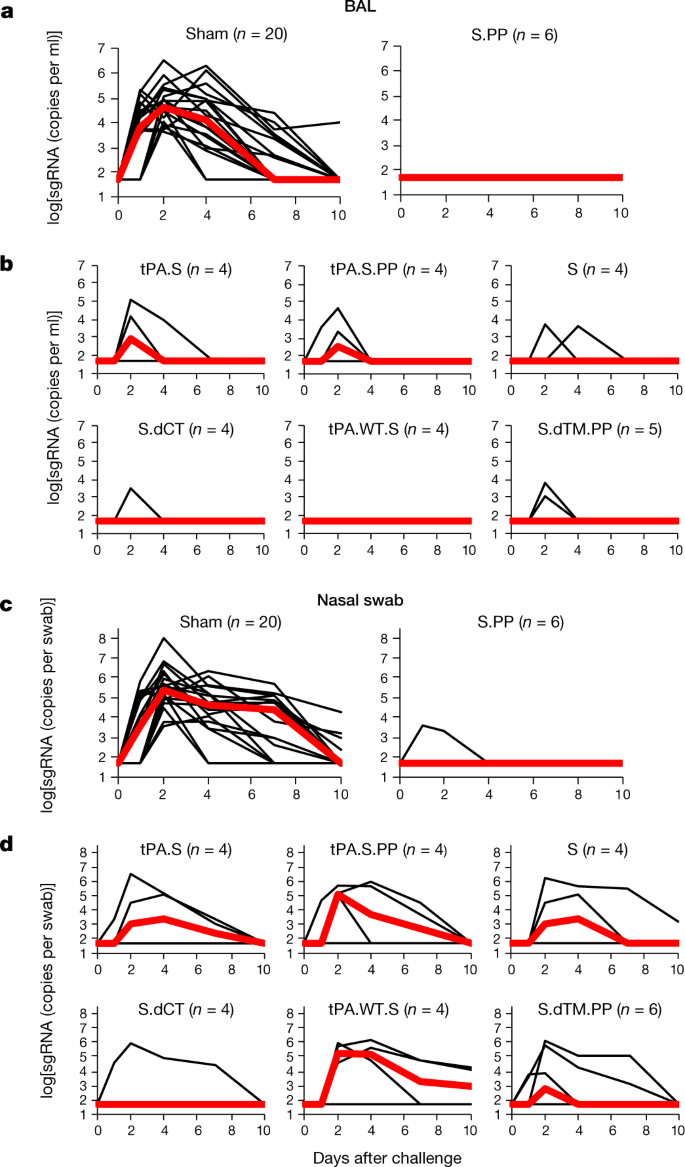
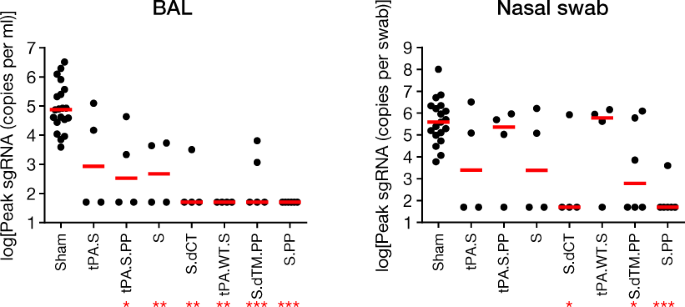
At week 6, all macaques were challenged with 1.0 × 105 50% tissue culture infectious dose (TCID50) of SARS-CoV-2 by the intranasal and intratracheal routes9,10. Consistent with previous observations, clinical disease was minimal in all macaques after challenge9,10. Viral loads in BAL and nasal swabs were assessed by reverse transcription PCR (RT–PCR) specific for subgenomic mRNA (sgRNA), which is thought to measure replicating virus9,20. All 20 sham controls were infected and showed a median peak of 4.89 (range 3.85–6.51) log10[sgRNA (copies per ml)] in BAL samples (Fig. 4a). By contrast, macaques that were treated with Ad26-S.PP had no detectable virus in BAL samples. Partial protection was observed with the other vaccines, with occasional macaques showing low levels of sgRNA in BAL (Fig. 4b). Similarly, sham controls showed a median peak of 5.59 (range 3.78–8.01) log10[sgRNA (copies per swab)] in nasal swabs (Fig. 4c). Only one of the macaques that received the Ad26-S.PP vaccine showed a low amount of virus in nasal swabs. The macaques that were treated with the other vaccines generally had reduced viral loads in nasal swabs compared with control macaques, although protection was optimal with Ad26-S.PP (Fig. 4d). All vaccinated macaques showed no detectable infectious virus in nasal swabs by plaque-forming unit (PFU) assays (Extended Data Fig. 5).
Fig. 4: Viral loads in rhesus macaques after SARS-CoV-2 challenge.
Rhesus macaques were challenged by the intranasal and intratracheal routes with 1.0 × 105 TCID50 of SARS-CoV-2. a, b, log10[sgRNA (copies per ml)] (limit of quantification 50 copies per ml) were assessed in BAL samples in sham controls and vaccinated macaques after challenge. c, d, log10[sgRNA (copies per swab)] (limit of quantification 50 copies per swab) were assessed in nasal swabs (NS) in sham controls and vaccinated macaques after challenge. Days after challenge are shown on the x axis. One macaque in the S.dTM.PP group did not have peak BAL samples obtained after challenge. Red lines reflect median values.
A comparison of peak viral loads in the vaccinated macaques suggested that protection in BAL samples was generally more robust than in nasal swabs (Fig. 5). The Ad26-S.PP vaccine provided complete protection in both the lower and upper respiratory tract with the exception of one macaque that showed a low amount of virus in nasal swabs, and resulted in greater than 3.2 and 3.9 log10-transformed reductions of median peak sgRNA in BAL and nasal swabs, respectively, as compared with sham controls (P < 0.0001 and P < 0.0001, respectively, two-sided Mann–Whitney tests) (Fig. 5). Among the 32 vaccinated macaques, 17 were completely protected and had no detectable sgRNA in BAL or nasal swabs after challenge, and 5 additional macaques had no sgRNA in BAL but showed some virus in nasal swabs.
Fig. 5: Summary of peak viral loads after SARS-CoV-2 challenge.
Peak viral loads in BAL and nasal swabs after challenge. Peak viral loads occurred variably on days 1–4 after challenge. Red lines reflect median viral loads. *P < 0.05, **P < 0.001, ***P < 0.0001, two-sided Mann–Whitney test. n = 4, 5 or 6 biologically independent macaques in vaccine groups and n = 20 biologically independent macaques in sham group.
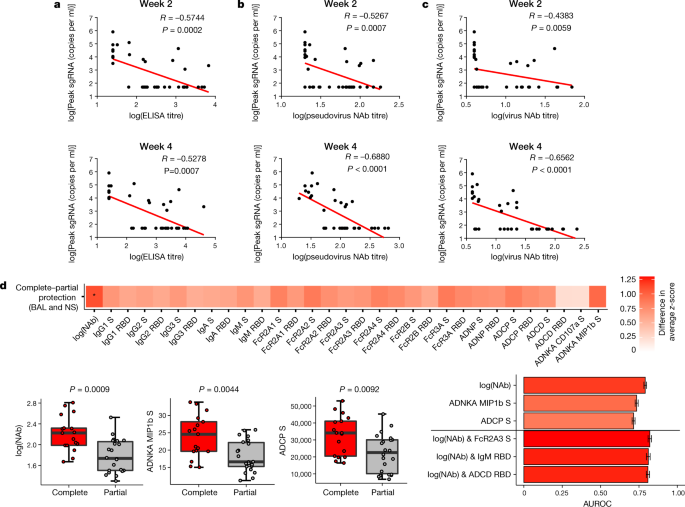
Immune correlates of protection
The size of this study and the variability in outcomes with the different vaccine constructs facilitated an immune correlates analysis. The log10(ELISA titre), log10(pseudovirus neutralizing antibody titre) and log10(live virus neutralizing antibody titre) at weeks 2 and 4 inversely correlated with peak log10(sgRNA) in both BAL (Fig. 6) and nasal swabs (Extended Data Fig. 6). In general, week-4 titres correlated better than week-2 titres, and neutralizing antibody titres correlated better than ELISA titres. The log10(pseudovirus neutralizing antibody titre) and log10(live virus neutralizing antibody titre) at week 4 inversely correlated with peak log10(sgRNA) in BAL (P < 0.0001, R = −0.6880 and P < 0.0001, R = −0.6562, respectively, two-sided Spearman rank-correlation test) (Fig. 6b, c) and in nasal swabs (P < 0.0001, R = −0.5839, and P < 0.0001, R = −0.5714, respectively, two-sided Spearman rank-correlation test) (Extended Data Fig. 6b, c). Together with previously published data10, these findings suggest that serum antibody titres may prove a useful immune correlate of protection for SARS-CoV-2 vaccines. By contrast, vaccine-elicited ELISPOT responses, and CD4+ and CD8+ intracellular cytokine staining responses, did not correlate with protection (data not shown).
Fig. 6: Antibody correlates of protection.
a–d, Correlations of binding ELISA titres (a), pseudovirus neutralizing antibody titres (b) and live virus neutralizing antibody titres (c) at weeks 2 and 4 with log[peak sgRNA (copies per ml)] in BAL after challenge. Red lines reflect the best linear fit relationship between these variables. P and R values reflect two-sided Spearman rank-correlation tests. n = 52 biologically independent macaques. d, The heat map shows the differences in the means of _z_-scored features between completely protected (n = 17) and partially protected and non-protected (n = 22) macaques. The two groups were compared by two-sided Mann–Whitney tests, and asterisks indicate the Benjamini–Hochberg corrected _q_-values (*q < 0.05), with q = 0.02707 for log(neutralizing antibodies). The dot plots show differences in the features that best discriminated completely protected and partially protected macaques, including neutralizing antibody titres, S-specific ADNKA and ADCP responses. P values were determined by two-sided Mann–Whitney tests. For the box plots, the upper bound of the box indicates the 75th percentile, and the lower bound the 25th percentile. The horizontal line shows the median, and the whiskers indicate minimum and maximum values. The bar plot shows the cross-validated area under the receiver operator characteristics curves using the features indicated on the x axis in a logistic regression model. The top three one-feature and two-feature models are shown. AUROC, areas under the receiver operator characteristics. Data are mean ± s.d. for 100 repetitions of tenfold cross-validation.
To gain further insight into antibody correlates of protection, we defined antibody parameters that distinguished completely protected macaques (defined as macaques with no detectable sgRNA in BAL or nasal swabs after challenge) and partially protected or non-protected macaques. The neutralizing antibody titre was the parameter most enriched in completely protected macaques compared with partially protected or non-protected macaques (P = 0.0009, two-sided Mann–Whitney test), followed by ADNKA (P = 0.0044) and ADCP (P = 0.0092) responses (Fig. 6d). Moreover, a logistic regression analysis showed that using two features, such as neutralizing antibody titre and FcγR2A-3, IgM or ADCD responses, improved correlation with protection (Fig. 6d). These data suggest that neutralizing antibodies are primarily responsible for protection against SARS-CoV-2 but that other binding and functional antibodies may also be involved.
Immune responses in vaccinated macaques after challenge
Sham controls and most of the vaccinated macaques (excluding Ad26-S.PP-vaccinated macaques) developed substantially higher pseudovirus neutralizing antibody responses (Extended Data Figs. 7, 8) as well as CD8+ and CD4+ T cell responses (Extended Data Fig. 9) by day 14 after SARS-CoV-2 challenge. CD8+ and CD4+ T cell responses were directed against several SARS-CoV-2 proteins, including spike (S1 and S2), nucleocapsid (NCAP), and non-structural (NS6, NS7a and NS8) proteins, in the sham controls. By contrast, macaques that were treated with the Ad26-S.PP vaccine did not show anamnestic neutralizing antibody responses (Extended Data Fig. 7) and only had low T cell responses against spike proteins (S1 and S2) (Extended Data Fig. 9), which was the vaccine antigen, after challenge. These findings are consistent with the largely undetectable viral loads in the Ad26-S.PP-vaccinated macaques (Figs. 4, 5) and suggest exceedingly low levels of viral replication in these macaques, if any at all, after challenge. Immunophenotyping of BAL cells from these macaques suggested similar cellular subpopulations in vaccinated and control macaques after immunization and challenge (Extended Data Fig. 10).
Discussion
The development of a safe and effective SARS-CoV-2 vaccine is a crucial global priority. Our data demonstrate that a single immunization with an Ad26 vector encoding a prefusion stabilized S immunogen (S.PP) induced robust neutralizing antibody responses and provided complete protection against SARS-CoV-2 challenge in five out of six rhesus macaques and near-complete protection in one out of six macaques. The S.PP immunogen contains the wild-type leader sequence, the full-length membrane-bound S protein, mutation of the furin cleavage site and two proline-stabilizing mutations15.
Our data extend recent preclinical studies of inactivated virus vaccines and DNA vaccines for SARS-CoV-2 in non-human primates10,21. Whereas inactivated virus vaccines and nucleic acid vaccines typically require two or more immunizations, some adenovirus vectors can induce robust and durable neutralizing antibody responses after a single immunization22,23,24. A single-shot SARS-CoV-2 vaccine would have important logistical and practical advantages compared with a two-dose vaccine for mass vaccination campaigns and control of the pandemic. However, we would expect that a two-dose vaccine with Ad26-S.PP would be more immunogenic. Our previous data demonstrate that a homologous boost with Ad26-HIV vectors augmented antibody titres by more than tenfold in both non-human primates and humans25,26,27, which suggests that both single-dose and two-dose regimens of the Ad26-S.PP vaccine should be evaluated in clinical trials. Baseline Ad26 neutralizing antibody titres in human populations11,28 have not suppressed the immunogenicity of an Ad26-HIV vaccine in several geographical regions25, suggesting the generalizability of this vector platform; however, this will be evaluated in clinical trials that are now underway.
Ad26-S.PP induced robust neutralizing antibody responses after a single immunization and provided complete protection against SARS-CoV-2 challenge in five out of six macaques, whereas one macaque had low levels of virus in nasal swabs. It is important to note that in the Ad26-S.PP-vaccinated macaques, neutralizing antibody titres and T cell responses did not expand after challenge, and T cell responses also did not broaden to non-vaccine antigens such as nucleocapsid and non-structural proteins. By contrast, sham controls and macaques that were treated with the other vaccines generated higher titres of neutralizing antibodies and T cell responses to several SARS-CoV-2 proteins after challenge, consistent with previous observations with DNA vaccines10. These data suggest minimal to no virus replication in the Ad26-S.PP-vaccinated macaques after SARS-CoV-2 challenge.
Vaccine-elicited titres of neutralizing antibodies before challenge correlated with protection in both BAL and nasal swabs after challenge, consistent with previous findings10. These data suggest that serum titres of neutralizing antibodies may be a potential biomarker for vaccine protection, although this will need to be confirmed in additional SARS-CoV-2 vaccine efficacy studies in both non-human primates and humans. Moreover, further functional antibody responses may also contribute to protection, such as ADNKA, ADCP and ADCD responses. The role of T cell responses in vaccine protection remains to be determined.
A limitation of our study is that we did not evaluate the durability of neutralizing antibody responses elicited by these vaccines, and future studies are planned to investigate this question. Additional studies could also evaluate mucosal delivery of this vaccine. Our studies were also not specifically designed to assess safety or the possibility of vaccine-associated enhanced respiratory disease or antibody-dependent enhancement of infection29. However, it is worth noting that the Ad26-S.PP vaccine elicited TH1-biased rather than TH2-biased T cell responses, and macaques with sub-protective neutralizing antibody titres did not demonstrate enhanced viral replication or clinical disease. Moreover, immunophenotyping of BAL cell subpopulations did not reveal increased eosinophils in vaccinated macaques compared with control macaques after immunization or challenge.
In summary, our data demonstrate that a single immunization of Ad26 vector-based vaccines for SARS-CoV-2 elicited robust neutralizing antibody titres and provided complete or near-complete protection against SARS-CoV-2 challenge in rhesus macaques. It is likely that protection in both the upper and lower respiratory tracts will be required to prevent transmission and disease in humans. The identification of a neutralizing antibody correlate of protection should prove useful in the clinical development of SARS-CoV-2 vaccines. The optimal Ad26-S.PP vaccine from this study, termed Ad26.COV2.S, is currently being evaluated in clinical trials.
Methods
No statistical methods were used to predetermine sample sizes. Cell lines were purchased from ATCC, and tested for mycoplasma.
Animals and study design
In total, 52 outbred Indian-origin adult male and female rhesus macaques (M. mulatta), 6–12 years old, were randomly allocated to groups. All macaques were housed at Bioqual. Macaques were treated with Ad26 vectors expressing tPA.S (n = 4), tPA.S.PP (n = 4), S (n = 4), S.dCT (n = 4), tPA.WT.S (n = 4), S.dTM.PP (n = 6) or S.PP (n = 6), and sham controls (n = 20). Macaques received a single immunization of 1011 viral particles of Ad26 vectors by the intramuscular route without adjuvant at week 0. At week 6, all macaques were challenged with 1.0 × 105 TCID50 (1.2 × 108 RNA copies, 1.1 × 104 PFU) SARS-CoV-2, which was derived from USA-WA1/2020 (NR-52281; BEI Resources)9. Viral particle titres were assessed by RT–PCR. Virus was administered as 1 ml by the intranasal route (0.5 ml in each nare) and 1 ml by the intratracheal route. All immunological and virological assays were performed blinded. All animal studies were conducted in compliance with all relevant local, state and federal regulations and were approved by the Bioqual Institutional Animal Care and Use Committee (IACUC).
Ad26 vectors
Ad26 vectors were constructed with seven variants of the SARS-CoV-2 spike (S) protein sequence (Wuhan/WIV04/2019; GenBank MN996528.1). Sequences were codon-optimized and synthesized. Replication-incompetent, E1/E3-deleted Ad26-vectors11 were produced in PER.C6.TetR cells using a plasmid containing the full Ad26 vector genome and a transgene expression cassette. Vectors were sequenced and tested for expression before use.
Western blot
For western blot analysis, 24-well plates were seeded with MRC-5 cells (1.25 × 105 cells per well), and after overnight growth they were transduced with Ad26 vectors encoding SARS-CoV-2 Spike transgenes. Cell lysates were collected 48 h after transduction and, after heating for 5 min at 85 °C, samples were loaded under non-reduced conditions on a precast 4–12% Bis-Tris SDS–PAGE gel (Invitrogen). Proteins were transferred to a nitrocellulose membrane using an iBlot2 dry blotting system (Invitrogen), and membrane blocking was performed overnight at 4 °C in Tris-buffered saline (TBS) containing 0.2% Tween 20 (v/v) (TBST) and 5% (w.v) Blotting-Grade Blocker (Bio-Rad). After overnight blocking, the membrane was incubated for 1 h with 2.8 μg ml−1 CR3046 in TBST-5% blocker. CR3046 is a human monoclonal antibody directed against SARS-CoV spike and binds to the spike S2 domain, and also cross-reacts with SARS-CoV-2 spike S2 (unpublished data). After incubation, the membrane was washed three times with TBST for 5 min and subsequently incubated for 1 h with 1:10,000 IRDye 800CW-conjugated goat-anti-human secondary antibody (Li-COR) in TBST-5% blocker. Finally, the PVDF membrane was washed three times with TBST for 5 min, and after drying developed using an ODYSSEY CLx Infrared Imaging System (Li-COR).
Subgenomic mRNA assay
SARS-CoV-2 E gene sgRNA was assessed by RT–PCR using primers and probes as previously described9,10,20. In brief, to generate a standard curve, the SARS-CoV-2 E gene sgRNA was cloned into a pcDNA3.1 expression plasmid; this insert was transcribed using an AmpliCap-Max T7 High Yield Message Maker Kit (Cellscript) to obtain RNA for standards. Before RT–PCR, samples collected from challenged macaques or standards were reverse-transcribed using Superscript III VILO (Invitrogen) according to the manufacturer’s instructions. A Taqman custom gene expression assay (ThermoFisher Scientific) was designed using the sequences targeting the E gene sgRNA20. Reactions were carried out on a QuantStudio 6 and 7 Flex Real-Time PCR System (Applied Biosystems) according to the manufacturer’s specifications. Standard curves were used to calculate sgRNA in copies per ml or per swab; the quantitative assay sensitivity was 50 copies per ml or per swab.
PFU assay
For plaque assays, confluent monolayers of Vero E6 cells (ATCC) were prepared in 6-well plates. Indicated samples collected from challenged macaques were serially diluted, added to wells, and incubated at 37 °C for 1 h. After incubation, 1.5 ml of 0.5% methylcellulose medium was added to each well and the plates were incubated at 37 °C with 5% CO2 for 2 days. Plates were fixed by adding 400 μl ice-cold methanol per well and incubating at −20 °C for 30 min. After fixation, the methanol was discarded, and cell monolayers were stained with 600 μl per well of 0.23% crystal violet for 30 min. After staining, the crystal violet was discarded, and the plates were washed once with 600 μl water to visualize and count plaques.
ELISA
RBD-specific binding antibodies were assessed by ELISA as previously described9,10. In brief, 96-well plates were coated with 1 μg ml−1 SARS-CoV-2 RBD protein (A. Schmidt, MassCPR) in 1× DPBS and incubated at 4 °C overnight. After incubation, plates were washed once with wash buffer (0.05% Tween 20 in 1× DPBS) and blocked with 350 μl casein block per well for 2–3 h at room temperature. After incubation, block solution was discarded and plates were blotted dry. Serial dilutions of heat-inactivated serum diluted in casein block were added to wells and plates were incubated for 1 h at room temperature, before three further washes and a 1-h incubation with a 1:1,000 dilution of anti-macaque IgG HRP (NIH NHP Reagent Program) at room temperature in the dark. Plates were then washed three times, and 100 μl of SeraCare KPL TMB SureBlue Start solution was added to each well; plate development was halted by the addition of 100 μl SeraCare KPL TMB Stop solution per well. The absorbance at 450 nm was recorded using a VersaMax or Omega microplate reader. ELISA endpoint titres were defined as the highest reciprocal serum dilution that yielded an absorbance >0.2. The log10(endpoint titres) are reported.
Pseudovirus neutralization assay
The SARS-CoV-2 pseudoviruses expressing a luciferase reporter gene were generated in an similar approach to that previously described9,10,16. In brief, the packaging construct psPAX2 (AIDS Resource and Reagent Program), luciferase reporter plasmid pLenti-CMV Puro-Luc (Addgene), and spike protein expressing pcDNA3.1-SARS-CoV-2 SΔCT were co-transfected into HEK293T cells with calcium phosphate. The supernatants containing the pseudotype viruses were collected 48 h after transfection; pseudotype viruses were purified by filtration with 0.45-μm filter. To determine the neutralization activity of the antisera from vaccinated macaques, HEK293T-hACE2 cells were seeded in 96-well tissue culture plates at a density of 1.75 × 104 cells per well overnight. Twofold serial dilutions of heat-inactivated serum samples were prepared and mixed with 50 μl of pseudovirus. The mixture was incubated at 37 °C for 1 h before adding to HEK293T-hACE2 cells. After 48 h, cells were lysed in Steady-Glo Luciferase Assay (Promega) according to the manufacturer’s instructions. SARS-CoV-2 neutralization titres were defined as the sample dilution at which a 50% reduction in relative light units was observed relative to the average of the virus control wells.
Live virus neutralization assay
A full-length SARS-CoV-2 virus based on the Seattle Washington isolate was designed to express nanoluciferase (nLuc) and GFP and was recovered via reverse genetics and previously described17,18. The SARS-CoV-2 nLuc-GFP virus titre was measured in Vero E6 USAMRIID cells, as defined by PFU per ml, in a 6-well plate format in quadruplicate biological replicates for accuracy. In addition, the virus was titred in Vero E6 USAMRID cells to ensure the relative light unit signal was at least ten times the cell only control background. For the 96-well neutralization assay, Vero E6 USAMRID cells were plated at 20,000 cells per well the day before in clear-bottom black-walled plates. Cells were inspected to ensure confluency on the day of assay. In separate 96-well dilution plates, neutralizing antibody serum samples were diluted to a starting dilution of 1:4 and were serially diluted fourfold up to eight dilution spots. Serially diluted serum samples were added in equal volume to 90 PFU of virus in duplicate test wells. Cell and virus-only control wells were also included in each 96-well dilution plate. The antibody–virus and virus-only mixtures were then incubated at 37 °C with 5% CO2 for exactly 1 h. After incubation, growth medium was removed from the clear-bottom black-walled 96-well plates and virus–antibody dilution complexes and virus-only and cell controls were added to the cells in duplicate and tips were replaced between each duplicate sample. After infection, 96-well neutralization assay plates were incubated at 37 °C with 5% CO2 for 48 h. After the 48-h incubation, cells were lysed, and luciferase activity was measured via Nano-Glo Luciferase Assay System (Promega) according to the manufacturer specifications. Luminescence was measured by a Spectramax M3 plate reader (Molecular Devices). SARS-CoV-2 neutralization titres were defined as the sample dilution at which a 50% reduction in relative light units was observed relative to the average of the virus control wells.
Systems serology
Luminex
A customized multiplexed approach to quantify relative antigen-specific antibody titres was used, as previously described30. Therefore, microspheres (Luminex) with a unique fluorescence were coupled with SARS-CoV-2 antigens including S protein and RBD via covalent _N_-hydroxysuccinimide (NHS)–ester linkages via EDC (Thermo Scientific) and Sulfo-NHS (Thermo Scientific). Per well in a 384-well plate (Greiner), 1.2 × 103 beads per region or antigen were added and incubated with diluted serum sample (1:100 for all isotypes or subclasses except for IgG1, which was diluted 1:250 as well as Fc-receptor binding) for 16 h shaking at 900 rpm at 4 °C. After formation of immune complexes, microspheres were washed three times in 0.1% BSA and 0.05% Tween 20 (Luminex assay buffer) with an automated plate washer (Tecan). Anti-rhesus IgG1, IgG2, IgG3, IgA (NIH NHP Reagent Program) and IgM (Life Diagnostic) detection antibodies were diluted in Luminex assay buffer to 0.65 μg ml−1 and incubated with beads for 1 h at room temperature while shaking at 900 rpm. After washing of stained immune complexes, a tertiary goat anti-mouse IgG-PE antibody (Southern Biotech) was added to each well at 0.5 μg ml−1 and incubated for 1 h at room temperature on a shaker. Similarly, for the Fc-receptor binding profiles, recombinant rhesus FcγR2A-1, FcγR2A-2, FcγR2A-3, FcγR2A-4, FcγR3A and human FcγR2B (Duke Protein Production facility) were biotinylated (Thermo Scientific) and conjugated to Streptavidin-PE for 10 min (Southern Biotech). The coated beads were then washed and read on a flow cytometer, iQue (Intellicyt) with a robot arm attached (PAA). Events were gated on each bead region, and the median fluorescence of phycoerythrin (PE) for bead-positive events was reported. Samples were run in duplicate per each secondary detection agent.
ADNP, ADCP and ADCD assays
ADNP, ADCP and ADCD assays were performed as previously described31,32,33. In brief, SARS-CoV-2 S and RBD were biotinylated (Thermo Fisher) and coupled to 1 μm yellow (ADCP and ADNP) and red (ADCD) fluorescent beads for 2 h at 37 °C. Excess antigen was removed by washing twice with 0.1% BSA in PBS. Next, 1.82 × 108 antigen-coated beads were added to each well of a 96-well plate and incubated with diluted samples (ADCP and ADNP 1:100, ADCD 1:10) at 37 °C for 2 h to facilitate immune complex formation. After the incubation, complexed beads were washed and for ADCP assays, 2.5 × 104 THP-1 cells (American Type Culture Collection) were added per well and incubated for 16 h at 37 °C. For ADNP assays, peripheral blood mononuclear cells (PBMCs) were isolated from healthy blood donors by lysis of red blood cells by addition of Ammonium-Chloride-Potassium (ACK) lysis (Thermo Fisher) and 5 × 104 cells were added per well and incubated for 1 h at 37 °C. Subsequently, primary blood cells were stained with an anti-Cd66b Pac blue detection antibody (BioLegend). For ADCD assays, lyophilized guinea pig complement was reconstituted according to manufacturer’s instructions (Cedarlane) with water and 4 μl per well were added in gelatin veronal buffer containing Mg2+ and Ca2+ (GVB++, Boston BioProducts) to the immune complexes for 20 min at 37 °C. After washing twice with 15 mM EDTA in PBS, immune complexes were stained with a fluorescein-conjugated goat IgG fraction to guinea pig complement C3 (MpBio). After incubation with THP-s and staining of cells for ADNP and ADCD cell samples are fixed with 4% paraformaldehyde and sample acquisition was performed via flow cytometry (Intellicyt, iQue Screener plus) using a robot arm (PAA). All events were gated on single cells and bead-positive events, for ADCP and ADNP assays, a phagocytosis score was calculated as the percentage of bead positive cells × GMFI/1,000; in which GMFI denotes geometric mean fluorescence intensity. For ADCD assays, the median of C3-positive events is reported. All samples were run in duplicate on separate days.
ADNKA assay
For analysis of natural killer cell-related responses, an ELISA-based assay was used. Therefore, 96-well ELISA plates (Thermo Fisher) were coated with SARS-CoV-2 S at 37 °C for 2 h. Plates were then washed and blocked with 5% BSA in PBS overnight at 4 °C. Natural killer cells were isolated from buffy coats from healthy donors (MGH blood donor centre) using the RosetteSep isolation kit (Stem Cell Technologies) and natural killer cells were rested overnight supplemented with IL-15 (Stemcell). Serum samples were diluted 1:50 and incubated at 37 °C for 2 h on the ELISA plates. A staining cocktail of anti-CD107a-PE-Cy5 stain (BD), brefeldin A (Sigma), and GolgiStop (BD) was added to the natural killer cells and 5 × 104 natural killer cells per well were added and incubated for 5 h at 37 °C. Natural killer cells were fixed and permeabilized using Perm A and B (Thermo Fisher) and surface markers were stained for with anti-CD16 APC-Cy7 (BD), anti-CD56 PE-Cy7 (BD) and anti-CD3 AlexaFluor 700 antibodies (BD). Intracellular staining included anti-IFNγ APC (BD) and anti-MIP-1β PE (BD). Acquisition occurred by flow cytometry iQue (Intellicyt), equipped with a robot arm (PAA). Natural killer cells were defined as CD3−, CD16+ and CD56+. The ADNKA assay was performed in duplicate across two blood donors.
Analysis
All isotypes or subclasses, Fc-receptor binding and ADCD data were log10-transformed. For the radar plots, each antibody feature was normalized such that its minimal value is 0 and the maximal value is 1 across groups before using the median within a group. A principal component analysis (PCA) was constructed using the R package ‘ropls’ to compare multivariate profiles. Completely protected macaques were defined as having no detectable sgRNA copies per ml in BAL and nasal swabs. Completely protected versus partially protected and non-protected macaques were compared using two-sided Mann–Whitney tests. For the visualization in the heat map, the differences in the means of completely and partially protected group of _z_-scored features were shown. To indicate significances in the heat map, a Benjamini–Hochberg correction was used to correct for multiple comparisons. To assess the ability of features and their combinations to predict protection, logistic regression models were trained for 100 repetitions in a tenfold cross-validation framework and areas under the receiver operator characteristics curves were calculated. All potential combinations of two features were tested. For this, the R packages glm and pROC were used.
IFNγ ELISPOT assay
ELISPOT plates were coated with mouse anti-human IFNγ monoclonal antibody from BD Pharmingen at a concentration of 5 μg per well overnight at 4 °C. Plates were washed with DPBS containing 0.25% Tween 20, and blocked with R10 medium (RPMI with 11% FBS and 1.1% penicillin-streptomycin) for 1 h at 37 °C. The spike 1 and spike 2 peptide pools contain 15 amino acid peptides overlapping by 11 amino acids that span the protein sequence and reflect the N- and C-terminal halves of the protein, respectively. Spike 1 and 2 peptide pools were prepared at a concentration of 2 μg per well, and 200,000 cells per well were added. The peptides and cells were incubated for 18–24 h at 37 °C. All steps following this incubation were performed at room temperature. The plates were washed with coulter buffer and incubated for 2 h with rabbit polyclonal anti-human IFNγ biotin from U-Cytech (1 μg ml−1). The plates are washed a second time and incubated for 2 h with Streptavidin-alkaline phosphatase antibody from Southern Biotechnology (1 μg ml−1). The final wash was followed by the addition of Nitor-blue Tetrazolium Chloride/5-bromo-4-chloro 3 ‘indolyl phosphate p-toludine salt (NBT/BCIP chromagen) substrate solution for 7 min. The chromagen was discarded and the plates were washed with water and dried in a dim place for 24 h. Plates were scanned and counted on a Cellular Technologies Limited Immunospot Analyzer.
IL-4 ELISPOT assay
Pre-coated monoclonal antibody IL-4 ELISPOT plates (Mabtech) were washed and blocked. The assay was then performed as described above except the development time with NBT/BCIP chromagen substrate solution was 12 min.
Intracellular cytokine staining assay
Approximately 106 PBMCs per well were re-suspended in 100 μl of R10 medium supplemented with CD49d monoclonal antibody (1 μg ml−1). Each sample was assessed with mock (100 μl of R10 plus 0.5% DMSO; background control), peptide pools (2 μg ml−1), or 10 pg ml−1 phorbol myristate acetate (PMA) and 1 μg ml−1 ionomycin (Sigma-Aldrich) (100 μl; positive control) and incubated at 37 °C for 1 h. After incubation, 0.25 μl of GolgiStop and 0.25 μl of GolgiPlug in 50 μl of R10 was added to each well and incubated at 37 °C for 8 h and then held at 4 °C overnight. The next day, the cells were washed twice with DPBS, stained with Near IR live/dead dye for 10 min and then stained with predetermined titres of monoclonal antibodies against CD279 (clone EH12.1, BB700), CD38 (clone OKT10, PE), CD28 (clone 28.2, PE CY5), CD4 (clone L200, BV510), CD45 (clone D058-1283, BUV615), CD95 (clone DX2, BUV737), CD8 (clone SK1, BUV805), for 30 min. Cells were then washed twice with 2% FBS/DPBS buffer and incubated for 15 min with 200 μkl of BD CytoFix/CytoPerm Fixation/Permeabilization solution. Cells were washed twice with 1× Perm Wash buffer (BD Perm/Wash Buffer 10× in the CytoFix/CytoPerm Fixation/ Permeabilization kit diluted with MilliQ water and passed through 0.22-μm filter) and stained with intracellularly with monoclonal antibodies against Ki67 (clone B56, FITC), CD69 (clone TP1.55.3, ECD), IL10 (clone JES3-9D7, PE CY7), IL-13 (clone JES10-5A2, BV421), TNF (clone Mab11, BV650), IL-4 (clone MP4-25D2, BV711), IFNγ (clone B27; BUV395), IL-2 (clone MQ1-17H12, APC), CD3 (clone SP34.2, Alexa 700), for 30 min. Cells were washed twice with 1× Perm Wash buffer and fixed with 250 μl of freshly prepared 1.5% formaldehyde. Fixed cells were transferred to 96-well round bottom plate and analysed by BD FACSymphony system.
Immunophenotyping of BAL cells
BAL cells were stained with Aqua live/dead dye for 20 min, washed with 2% FBS/DPBS buffer, and stained with monoclonal antibodies against CD8 (clone SK1, FITC), CD123 (clone 7G3, PE), CD28 (clone 28.2, PE CF594), CD4 (clone L200, BB700), CD159a (clone Z199, PE CY7), CD49d (clone 9F10, BV421), CD20 (clone 2H7, BV570), CD45 (clone D058-1283, BV605), TCR γδ (clone B1, BV650), CD95 (clone DX2, BV711), CD163 (clone GHI/61, BV786), CD16 (clone 3G8, BUV395), CD14 (clone M5E2, BUV737), CD66 (clone TET2, APC), CD3 (clone SP34.2, Alexa 700), HLA-DR (clone G46-6, APC H7), for 30 min. After staining, cells were washed twice with 2% FBS/DPBS buffer and fixed by 1.5% formaldehyde. All data were acquired on a BD LSRII flow cytometer with FACSDiva software (BD Biosciences). Subsequent analyses were performed using FlowJo software (Treestar, v.9.9.6). For subpopulation quantification, dead cells were excluded by Aqua dye and CD45 was used as a positive inclusion gate for all leukocytes. Lymphocyte populations were quantified using standard rhesus macaque phenotyping protocols and macrophage/granulocyte populations were identified among high granular fractions: macrophages (CD163+); eosinophils (CD66abce+CD49dhi/modCD14−); neutrophils (CD66abce+CD49dmodCD14+); basophils CD66abce−HLA−DR−CD123hi).
Statistical analyses
Analysis of virological and immunologic data was performed using GraphPad Prism 8.4.2 (GraphPad Software). Comparison of data between groups was performed using two-sided Mann–Whitney tests. Correlations were assessed by two-sided Spearman rank-correlation tests. P values of less than 0.05 were considered significant.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
All data are available in the Article and its Supplementary Information. Source data are provided with this paper.
Change history
19 January 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41586-020-03100-y
References
- Wu, F. et al. A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020).
ADS CAS PubMed PubMed Central Google Scholar - Zhou, P. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020).
Article ADS CAS PubMed PubMed Central Google Scholar - Holshue, M. L. et al. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 382, 929–936 (2020).
Article CAS PubMed PubMed Central Google Scholar - Li, Q. et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 382, 1199–1207 (2020).
Article CAS PubMed PubMed Central Google Scholar - Zhu, N. et al. A Novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733 (2020).
Article CAS PubMed Google Scholar - Chen, N. et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 395, 507–513 (2020).
Article CAS PubMed PubMed Central Google Scholar - Huang, C. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020).
Article CAS PubMed PubMed Central Google Scholar - Chan, J. F. et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 395, 514–523 (2020).
Article CAS PubMed PubMed Central Google Scholar - Chandrashekar, A. et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science 369, 812–817 (2020).
Article CAS PubMed PubMed Central Google Scholar - Yu, J. et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 369, 806–811 (2020).
Article CAS PubMed PubMed Central Google Scholar - Abbink, P. et al. Comparative seroprevalence and immunogenicity of six rare serotype recombinant adenovirus vaccine vectors from subgroups B and D. J. Virol. 81, 4654–4663 (2007).
Article CAS PubMed PubMed Central Google Scholar - Alharbi, N. K. et al. ChAdOx1 and MVA based vaccine candidates against MERS-CoV elicit neutralising antibodies and cellular immune responses in mice. Vaccine 35, 3780–3788 (2017).
Article CAS PubMed PubMed Central Google Scholar - Kirchdoerfer, R. N. et al. Pre-fusion structure of a human coronavirus spike protein. Nature 531, 118–121 (2016).
Article ADS CAS PubMed PubMed Central Google Scholar - Pallesen, J. et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl Acad. Sci. USA 114, E7348–E7357 (2017).
Article CAS PubMed PubMed Central Google Scholar - Wrapp, D. et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020).
Article ADS CAS PubMed PubMed Central Google Scholar - Yang, Z. Y. et al. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature 428, 561–564 (2004).
Article ADS CAS PubMed PubMed Central Google Scholar - Scobey, T. et al. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc. Natl Acad. Sci. USA 110, 16157–16162 (2013).
Article ADS CAS PubMed PubMed Central Google Scholar - Yount, B. et al. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl Acad. Sci. USA 100, 12995–13000 (2003).
Article ADS CAS PubMed PubMed Central Google Scholar - Chung, A. W. et al. Dissecting polyclonal vaccine-induced humoral immunity against HIV using systems serology. Cell 163, 988–998 (2015).
Article CAS PubMed PubMed Central Google Scholar - Wölfel, R. et al. Virological assessment of hospitalized patients with COVID-2019. Nature 581, 465–469 (2020).
Article ADS PubMed Google Scholar - Gao, Q. et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science 369, 77–81 (2020).
Article ADS CAS PubMed Google Scholar - Abbink, P. et al. Durability and correlates of vaccine protection against Zika virus in rhesus monkeys. Sci. Transl. Med. 9, eaao4163 (2017).
Article PubMed PubMed Central Google Scholar - Abbink, P. et al. Protective efficacy of multiple vaccine platforms against Zika virus challenge in rhesus monkeys. Science 353, 1129–1132 (2016).
Article ADS CAS PubMed PubMed Central Google Scholar - Cox, F. et al. Adenoviral vector type 26 encoding Zika virus (ZIKV) M-Env antigen induces humoral and cellular immune responses and protects mice and nonhuman primates against ZIKV challenge. PLoS ONE 13, e0202820 (2018).
Article PubMed PubMed Central Google Scholar - Barouch, D. H. et al. Evaluation of a mosaic HIV-1 vaccine in a multicentre, randomised, double-blind, placebo-controlled, phase 1/2a clinical trial (APPROACH) and in rhesus monkeys (NHP 13-19). Lancet 392, 232–243 (2018).
Article CAS PubMed PubMed Central Google Scholar - Barouch, D. H. et al. Protective efficacy of adenovirus/protein vaccines against SIV challenges in rhesus monkeys. Science 349, 320–324 (2015).
Article ADS CAS PubMed PubMed Central Google Scholar - Baden, L. R. et al. First-in-human evaluation of the safety and immunogenicity of a recombinant adenovirus serotype 26 HIV-1 Env vaccine (IPCAVD 001). J. Infect. Dis. 207, 240–247 (2013).
Article CAS PubMed Google Scholar - Barouch, D. H. et al. International seroepidemiology of adenovirus serotypes 5, 26, 35, and 48 in pediatric and adult populations. Vaccine 29, 5203–5209 (2011).
Article CAS PubMed PubMed Central Google Scholar - Graham, B. S. Rapid COVID-19 vaccine development. Science 368, 945–946 (2020).
Article CAS PubMed Google Scholar - Brown, E. P. et al. Multiplexed Fc array for evaluation of antigen-specific antibody effector profiles. J. Immunol. Methods 443, 33–44 (2017).
Article CAS PubMed PubMed Central Google Scholar - Ackerman, M. E. et al. A robust, high-throughput assay to determine the phagocytic activity of clinical antibody samples. J. Immunol. Methods 366, 8–19 (2011).
Article CAS PubMed Google Scholar - Lu, L. L. et al. A functional role for antibodies in tuberculosis. Cell 167, 433–443 (2016).
Article CAS PubMed PubMed Central Google Scholar - Fischinger, S. et al. A high-throughput, bead-based, antigen-specific assay to assess the ability of antibodies to induce complement activation. J. Immunol. Methods 473, 112630 (2019).
Article CAS PubMed PubMed Central Google Scholar
Acknowledgements
We thank S. Blokland, Y. Choi, K. de Boer, I. de los Rios Oakes, E. van der Helm, D. Spek, I. Swart, M. Koning, A. Brandjes, N. van Dijk, A. de Wilde, M. Navis, R. van Schie, J. Verhagen, R. Vogels, R. van der Vlugt, A. Roos Broekhuijsen, B. Bart, J. Velasco, B. Finneyfrock, C. Shaver, J. Yalley-Ogunro, D. Wesemann, N. Kordana, M. Lifton, E. Borducchi, M. Silva, A. Richardson and C. Caron for advice, assistance and reagents. This project was funded in part by the Department of Health and Human Services Biomedical Advanced Research and Development Authority (BARDA) under contract HHS0100201700018C. We also acknowledge support from Janssen Vaccines & Prevention BV, the Ragon Institute of MGH, MIT, and Harvard, Mark and Lisa Schwartz Foundation, Massachusetts Consortium on Pathogen Readiness (MassCPR), and the National Institutes of Health (OD024917, AI129797, AI124377, AI128751, AI126603 to D.H.B.; AI007151 and AI152296 to D.R.M.; AI146779 to A.G.S.; 272201700036I-0-759301900131-1, AI100625, AI110700, AI132178, AI149644, AI108197 to R.S.B.). We also acknowledge a Burroughs Wellcome Fund Postdoctoral Enrichment Program Award to D.R.M.
Author information
Author notes
- These authors contributed equally: Noe B. Mercado, Roland Zahn, Frank Wegmann, Carolin Loos, Abishek Chandrashekar, Jingyou Yu, Jinyan Liu, Lauren Peter, Katherine McMahan, Lisa H. Tostanoski, Xuan He, David R. Martinez
- These authors jointly supervised this work: Hanneke Schuitemaker, Dan H. Barouch
Authors and Affiliations
- Center for Virology and Vaccine Research, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA, USA
Noe B. Mercado, Abishek Chandrashekar, Jingyou Yu, Jinyan Liu, Lauren Peter, Katherine McMahan, Lisa H. Tostanoski, Xuan He, Esther A. Bondzie, Gabriel Dagotto, Makda S. Gebre, Emily Hoffman, Catherine Jacob-Dolan, Marinela Kirilova, Zhenfeng Li, Zijin Lin, Shant H. Mahrokhian, Lori F. Maxfield, Felix Nampanya, Ramya Nityanandam, Joseph P. Nkolola, Shivani Patel, John D. Ventura, Kaylee Verrington, Huahua Wan, R. Keith Reeves & Dan H. Barouch - Janssen Vaccines and Prevention BV, Leiden, The Netherlands
Roland Zahn, Frank Wegmann, Lucy Rutten, Rinke Bos, Danielle van Manen, Jort Vellinga, Jerome Custers, Johannes P. Langedijk, Ted Kwaks, Mark J. G. Bakkers, David Zuijdgeest, Sietske K. Rosendahl Huber, Paul Stoffels, Mathai Mammen, Johan Van Hoof & Hanneke Schuitemaker - Ragon Institute of MGH, MIT and Harvard, Cambridge, MA, USA
Carolin Loos, Caroline Atyeo, Stephanie Fischinger, John S. Burke, Jared Feldman, Blake M. Hauser, Timothy M. Caradonna, Aaron G. Schmidt, Galit Alter & Dan H. Barouch - Massachusetts Institute of Technology, Cambridge, MA, USA
Carolin Loos & Douglas A. Lauffenburger - University of North Carolina at Chapel Hill, Chapel Hill, NC, USA
David R. Martinez & Ralph S. Baric - Harvard Medical School, Boston, MA, USA
Caroline Atyeo, Stephanie Fischinger, Jared Feldman, Blake M. Hauser, Timothy M. Caradonna, Gabriel Dagotto, Makda S. Gebre, Catherine Jacob-Dolan, Aaron G. Schmidt & Dan H. Barouch - Bioqual, Rockville, MD, USA
Laurent Pessaint, Alex Van Ry, Kelvin Blade, Amanda Strasbaugh, Mehtap Cabus, Renita Brown, Anthony Cook, Serge Zouantchangadou, Elyse Teow, Hanne Andersen & Mark G. Lewis - Children’s Hospital, Boston, MA, USA
Yongfei Cai & Bing Chen - Massachusetts Consortium on Pathogen Readiness, Boston, MA, USA
Bing Chen, Aaron G. Schmidt, Galit Alter & Dan H. Barouch
Authors
- Noe B. Mercado
You can also search for this author inPubMed Google Scholar - Roland Zahn
You can also search for this author inPubMed Google Scholar - Frank Wegmann
You can also search for this author inPubMed Google Scholar - Carolin Loos
You can also search for this author inPubMed Google Scholar - Abishek Chandrashekar
You can also search for this author inPubMed Google Scholar - Jingyou Yu
You can also search for this author inPubMed Google Scholar - Jinyan Liu
You can also search for this author inPubMed Google Scholar - Lauren Peter
You can also search for this author inPubMed Google Scholar - Katherine McMahan
You can also search for this author inPubMed Google Scholar - Lisa H. Tostanoski
You can also search for this author inPubMed Google Scholar - Xuan He
You can also search for this author inPubMed Google Scholar - David R. Martinez
You can also search for this author inPubMed Google Scholar - Lucy Rutten
You can also search for this author inPubMed Google Scholar - Rinke Bos
You can also search for this author inPubMed Google Scholar - Danielle van Manen
You can also search for this author inPubMed Google Scholar - Jort Vellinga
You can also search for this author inPubMed Google Scholar - Jerome Custers
You can also search for this author inPubMed Google Scholar - Johannes P. Langedijk
You can also search for this author inPubMed Google Scholar - Ted Kwaks
You can also search for this author inPubMed Google Scholar - Mark J. G. Bakkers
You can also search for this author inPubMed Google Scholar - David Zuijdgeest
You can also search for this author inPubMed Google Scholar - Sietske K. Rosendahl Huber
You can also search for this author inPubMed Google Scholar - Caroline Atyeo
You can also search for this author inPubMed Google Scholar - Stephanie Fischinger
You can also search for this author inPubMed Google Scholar - John S. Burke
You can also search for this author inPubMed Google Scholar - Jared Feldman
You can also search for this author inPubMed Google Scholar - Blake M. Hauser
You can also search for this author inPubMed Google Scholar - Timothy M. Caradonna
You can also search for this author inPubMed Google Scholar - Esther A. Bondzie
You can also search for this author inPubMed Google Scholar - Gabriel Dagotto
You can also search for this author inPubMed Google Scholar - Makda S. Gebre
You can also search for this author inPubMed Google Scholar - Emily Hoffman
You can also search for this author inPubMed Google Scholar - Catherine Jacob-Dolan
You can also search for this author inPubMed Google Scholar - Marinela Kirilova
You can also search for this author inPubMed Google Scholar - Zhenfeng Li
You can also search for this author inPubMed Google Scholar - Zijin Lin
You can also search for this author inPubMed Google Scholar - Shant H. Mahrokhian
You can also search for this author inPubMed Google Scholar - Lori F. Maxfield
You can also search for this author inPubMed Google Scholar - Felix Nampanya
You can also search for this author inPubMed Google Scholar - Ramya Nityanandam
You can also search for this author inPubMed Google Scholar - Joseph P. Nkolola
You can also search for this author inPubMed Google Scholar - Shivani Patel
You can also search for this author inPubMed Google Scholar - John D. Ventura
You can also search for this author inPubMed Google Scholar - Kaylee Verrington
You can also search for this author inPubMed Google Scholar - Huahua Wan
You can also search for this author inPubMed Google Scholar - Laurent Pessaint
You can also search for this author inPubMed Google Scholar - Alex Van Ry
You can also search for this author inPubMed Google Scholar - Kelvin Blade
You can also search for this author inPubMed Google Scholar - Amanda Strasbaugh
You can also search for this author inPubMed Google Scholar - Mehtap Cabus
You can also search for this author inPubMed Google Scholar - Renita Brown
You can also search for this author inPubMed Google Scholar - Anthony Cook
You can also search for this author inPubMed Google Scholar - Serge Zouantchangadou
You can also search for this author inPubMed Google Scholar - Elyse Teow
You can also search for this author inPubMed Google Scholar - Hanne Andersen
You can also search for this author inPubMed Google Scholar - Mark G. Lewis
You can also search for this author inPubMed Google Scholar - Yongfei Cai
You can also search for this author inPubMed Google Scholar - Bing Chen
You can also search for this author inPubMed Google Scholar - Aaron G. Schmidt
You can also search for this author inPubMed Google Scholar - R. Keith Reeves
You can also search for this author inPubMed Google Scholar - Ralph S. Baric
You can also search for this author inPubMed Google Scholar - Douglas A. Lauffenburger
You can also search for this author inPubMed Google Scholar - Galit Alter
You can also search for this author inPubMed Google Scholar - Paul Stoffels
You can also search for this author inPubMed Google Scholar - Mathai Mammen
You can also search for this author inPubMed Google Scholar - Johan Van Hoof
You can also search for this author inPubMed Google Scholar - Hanneke Schuitemaker
You can also search for this author inPubMed Google Scholar - Dan H. Barouch
You can also search for this author inPubMed Google Scholar
Contributions
D.H.B., R.Z., F.W., P.S., M.M., J.V.H. and H.S. designed the study and reviewed all data. R.Z., F.W., L.R., R.B., D.M., J.V., J.C., J.P.L., T.K., M.J.G.B., D.Z., S.K.R.H., H.S., B.C., J.L., Z.L. and D.H.B. designed the vaccines. N.B.M., A.C., J.Y., J.L., L.P., K.M., L.H.T., E.A.B., G.D., M.S.G., X.H., E.H., C.J.D., M.K., Z.L., S.H.M., L.F.M., F.N., R.N., J.P.N., S.P., J.D.V., K.V., H.W. and R.K.R. performed the immunological and virological assays. C.L., C.A., S.F., J.S.B., D.A.L. and G.A. performed the systems serology. D.R.M. and R.S.B. performed the live virus neutralization assays. L.P., A.V.R., K.B., A.S., M.C., R.B., A.C., S.Z., E.T., H.A. and M.G.L. led the clinical care of the macaques. J.F., B.M.H., T.M.C., Y.C., B.C. and A.G.S. provided purified proteins. D.H.B. wrote the paper with all co-authors.
Corresponding author
Correspondence toDan H. Barouch.
Ethics declarations
Competing interests
D.H.B., R.Z., F.W., L.R., R.B., D.M., J.V., J.C., J.P.L., T.K., M.J.G.B. and H.S are co-inventors on provisional vaccine patents (62/969,008; 62/994,630). R.Z., F.W., L.R., R.B., D.M., J.V., J.C., J.P.L., T.K., M.J.G.B., D.Z., S.K.R.H., P.S., M.M., J.V.H. and H.S. are employees of Janssen Vaccines and Prevention BV and hold stock in Johnson and Johnson.
Additional information
Peer review information Nature thanks Wolfgang Baumgärtner, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Correlation of pseudovirus neutralizing antibody titres and ELISA or live virus neutralizing antibody assays in vaccinated macaques.
Red line reflects the best linear fit relationship between these variables. P and R values reflect two-sided Spearman rank-correlation tests. n = 38 biologically independent macaques.
Extended Data Fig. 2 Peripheral and mucosal humoral immune responses in vaccinated rhesus macaques.
a, Comparison of pseudovirus neutralizing antibodies in macaques vaccinated with Ad26-S.PP (n = 6 biologically independent macaques) with previously reported cohorts of convalescent macaques9 (n = 9 biologically independent macaques) and convalescent humans10 (n = 27 biologically independent humans) who had recovered from SARS-CoV-2 infection. NHP, non-human primate. The upper bound of the box is the seventy-fifth and the lower bound the twenty-fifth percentile, the horizontal line indicates the median and the whiskers extend from the box bounds to the minimum/maximum value. b, S-specific IgG and IgA at week 4 in BAL by ELISA in sham controls and in Ad26-S.PP vaccinated macaques. Red bars reflect median responses.
Extended Data Fig. 3 Systems serology in vaccinated rhesus macaques.
a, S-specific and RBD-specific ADNP, ADCP, ADCD and ADNKA assays are shown. Red bars reflect median responses. b, S-specific and RBD-specific ADNP, ADCP, ADCD and ADNKA assays at week 4 are shown as radar plots. The size and colour intensity of the wedges indicate the median of the feature for the corresponding group (blue depicts antibody functions; red depicts antibody isotype, subclass or FcγR binding). The principal component analysis (PCA) plot shows the multivariate antibody profiles across groups. The PCA analysis reduces the dimension of the data by finding principal components, which are linear combinations of the original antibody features that are uncorrelated and best capture the variance in the data. Here, PC1 explains 74.2% and PC2 explains 8.3% of the variance. Each dot represents a macaque, the colour of the dot denotes the group, and the ellipses shows the distribution of the groups as 70% confidence levels assuming a multivariate normal distribution.
Extended Data Fig. 4 Cellular immune responses in vaccinated rhesus macaques.
IFNγ and IL-4 ELISPOT responses in response to pooled S peptides were assessed in a separate cohort of 10 macaques that received 1 × 1011 viral particles of the Ad26-S.PP vaccine at week 2 after vaccination. Red bars reflect median responses. Dotted lines reflect assay limit of quantification.
Extended Data Fig. 5 Infectious virus after challenge.
Infectious virus titres were assessed by PFU assays in nasal swabs on day 2 after challenge in vaccinated macaques and additional sham controls.
Extended Data Fig. 6 Correlates of protection in nasal swabs.
a–c, Correlations of binding ELISA titres (a), pseudovirus neutralizing antibody titres (b), and (c) live virus neutralizing antibody titres at weeks 2 and 4 with log[peak sgRNA (copies per swab)] in nasal swabs after challenge. Red lines reflect the best linear fit relationship between these variables. P and R values reflect two-sided Spearman rank-correlation tests. n = 52 biologically independent macaques.
Extended Data Fig. 7 Neutralizing antibody titres after SARS-CoV-2 challenge.
Pseudovirus neutralizing antibody titres before challenge and on day 14 after challenge in sham controls and in Ad26-S.PP vaccinated macaques. Red bars reflect median responses. Dotted lines reflect assay limit of quantification.
Extended Data Fig. 8 Neutralizing antibody titres after SARS-CoV-2 challenge.
Pseudovirus neutralizing antibody titres before challenge and on day 14 after challenge in vaccinated macaques. Red bars reflect median responses. Dotted lines reflect assay limit of quantification.
Extended Data Fig. 9 Cellular immune responses after SARS-CoV-2 challenge.
IFNγ+CD8+ and IFNγ+CD4+ T cell responses by intracellular cytokine staining assays in response to pooled spike (S1 and S2), nucleocapsid (NCAP), and non-structural proteins (N6, N7a and N8) peptides on day 14 after challenge in sham controls and in Ad26-S.PP-vaccinated macaques. Red bars reflect median responses. Dotted lines reflect assay limit of quantification.
Extended Data Fig. 10 Immunophenotyping of BAL cell subpopulations.
BAL cells from Ad26-S.PP-vaccinated and control macaques from 2 weeks after immunization and 2 weeks after challenge were assessed by flow cytometry for cellular subpopulations.
Supplementary information
Source data
Rights and permissions
About this article
Cite this article
Mercado, N.B., Zahn, R., Wegmann, F. et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques.Nature 586, 583–588 (2020). https://doi.org/10.1038/s41586-020-2607-z
- Received: 20 June 2020
- Accepted: 24 July 2020
- Published: 30 July 2020
- Issue Date: 22 October 2020
- DOI: https://doi.org/10.1038/s41586-020-2607-z