ImJoy: an open-source computational platform for the deep learning era (original) (raw)
- Correspondence
- Published: 28 November 2019
Nature Methods volume 16, pages 1199–1200 (2019)Cite this article
- 9416 Accesses
- 135 Citations
- 43 Altmetric
- Metrics details
Subjects
To the Editor — Deep learning (DL) methods achieve breakthrough performances in analyzing biomedical data across countless tasks, including medical diagnostics, DNA sequence analysis, augmented microscopy and drug design. Combined with increasing data repositories in genomics, imaging and other fields, such successes underlay a growing demand to adapt DL methods to new datasets and questions1. However, the dissemination of DL approaches faces considerable hurdles. Most published DL studies2,3,4,5 require users to retrain models on their own data to obtain the best performance and/or avoid erroneous results. Although trained models are frequently available through web applications or ImageJ plugins, retraining is typically only possible via scripts or command lines, rather than graphical user interfaces (GUIs). In addition, the complexities of setting up the required hardware and software environments often constitute forbidding obstacles[6](/articles/s41592-019-0627-0#ref-CR6 "Moen, E. et al. Nat. Methods https://doi.org/10.1038/s41592-019-0403-1
(2019)."). Furthermore, the large datasets and computational resources typical of current DL successes pose challenges to traditional desktop-oriented software that tightly couple GUI and computation. Cloud services can partly alleviate these difficulties, but raise privacy and confidentiality issues that can be prohibitive for medical data7. Meanwhile, deploying scientific software to mobile platforms can make them accessible to billions of people8, enabling large-scale biomedical research and citizen science. These opportunities and challenges call for new computational frameworks.
By leveraging various web libraries, such as three.js and D3, ImJoy allows users to build rich and interactive applications (Fig. 1a). For example, the ImageAnnotator plugin allows annotation of images, a pre-requisite for training segmentation methods, and can run on touchscreen devices (for example, smartphones). The HPA-UMAP plugin visualizes protein localization features computed by DL from Human Protein Atlas images9. Such interactive visualizations are instrumental to exploring massive databases and interacting with cloud services.
This is a preview of subscription content, access via your institution
Relevant articles
Open Access articles citing this article.
High-throughput image processing software for the study of nuclear architecture and gene expression
- Adib Keikhosravi
- , Faisal Almansour
- … Gianluca Pegoraro
Scientific Reports Open Access 08 August 2024
A low-cost smartphone fluorescence microscope for research, life science education, and STEM outreach
- Madison A. Schaefer
- , Heather N. Nelson
- … Jacob H. Hines
Scientific Reports Open Access 09 March 2023
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Additional access options:
Fig. 1: Overview of ImJoy.
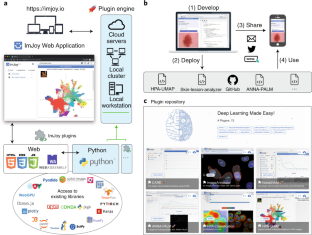
Code Availability
Source code for ImJoy and the example plugins is available at https://imjoy.io and https://github.com/imjoy-team/example-plugins, respectively.
References
- Ouyang, W. & Zimmer, C. Curr. Opin. Syst. Biol. 4, 105–113 (2017).
Article Google Scholar - Falk, T. et al. Nat. Methods 16, 67 (2019).
Article CAS Google Scholar - Alipanahi, B., Delong, A., Weirauch, M. T. & Frey, B. J. Nat. Biotechnol. 33, 831–838 (2015).
Article CAS Google Scholar - Ouyang, W., Aristov, A., Lelek, M., Hao, X. & Zimmer, C. Nat. Biotechnol. 36, 460–468 (2018).
Article CAS Google Scholar - Weigert, M. et al. Nat. Methods 15, 1090–1097 (2018).
Article CAS Google Scholar - Moen, E. et al. Nat. Methods https://doi.org/10.1038/s41592-019-0403-1 (2019).
- Tang, H. et al. BMC Med. Genomics 9, 63 (2016).
Article Google Scholar - Esteva, A. et al. Nature 542, 115–118 (2017).
Article CAS Google Scholar - Thul, P. J. et al. Science 356, eaal3321 (2017).
Article Google Scholar - Lee, H. et al. Nat. Biomed. Eng. 3, 173–182 (2019).
Article Google Scholar
Acknowledgements
This work was funded by the Institut Pasteur. W.O. was a scholar in the Pasteur–Paris University (PPU) International PhD program and was partly funded by a Fondation de la Recherche Médicale (FRM) grant to C.Z. (DEQ 20150331762). W.O. is a postdoctoral researcher supported by the Knut and Alice Wallenberg Foundation (2016.0204) and Erling-Persson Foundation (20180316) grants to E.L. We also acknowledge Investissement d’Avenir grant ANR-16-CONV-0005 for funding a GPU farm used for testing ImJoy. We thank the IT department of Institut Pasteur, in particular S. Fournier and T. Menard, for providing access to the kubernetes cluster and DGX-1 server for running and testing the ImJoy plugin engine and for technical support. We thank Q.T. Huynh for maintaining the GPU farm and for advice and assistance during the development of ImJoy. We also thank A. Martinez Casals, P. Thul, H. Xu, A. Aristov, A. Cesnik, C. Gnann, J. Parmar, K.M. Douglass, N. Stuurman, X. Hao, S. Dai, A. Hu, D. Guo, K. Zhou for testing and helping with ImJoy plugin development. We thank E. Rensen for proofreading the manuscript. We thank J. Nunez-Iglesias, S. Mehta, B. Chhun, J. Batson, L. Royer, N. Sofroniew and M. Woringer for useful advice and discussion.
Author information
Authors and Affiliations
- Imaging and Modeling Unit, Institut Pasteur, UMR 3691 CNRS, C3BI USR 3756 IP CNRS, Paris, France
Wei Ouyang, Florian Mueller & Christophe Zimmer - Science for Life Laboratory, School of Engineering Sciences in Chemistry, Biotechnology and Health, KTH - Royal Institute of Technology, Stockholm, Sweden
Wei Ouyang, Martin Hjelmare & Emma Lundberg - Department of Genetics, Stanford University, Stanford, CA, USA
Emma Lundberg - Chan Zuckerberg Biohub, San Francisco, CA, USA
Emma Lundberg
Authors
- Wei Ouyang
- Florian Mueller
- Martin Hjelmare
- Emma Lundberg
- Christophe Zimmer
Corresponding authors
Correspondence toWei Ouyang or Florian Mueller.
Supplementary Information
Rights and permissions
About this article
Cite this article
Ouyang, W., Mueller, F., Hjelmare, M. et al. ImJoy: an open-source computational platform for the deep learning era.Nat Methods 16, 1199–1200 (2019). https://doi.org/10.1038/s41592-019-0627-0
- Published: 28 November 2019
- Issue Date: December 2019
- DOI: https://doi.org/10.1038/s41592-019-0627-0
This article is cited by
BiaPy: accessible deep learning on bioimages
- Daniel Franco-Barranco
- Jesús A. Andrés-San Román
- Ignacio Arganda-Carreras
Nature Methods (2025)
Arkitekt: streaming analysis and real-time workflows for microscopy
- Johannes Roos
- Stéphane Bancelin
- Jean-Baptiste Sibarita
Nature Methods (2024)
JDLL: a library to run deep learning models on Java bioimage informatics platforms
- Carlos García López de Haro
- Stéphane Dallongeville
- Jean-Christophe Olivo-Marin
Nature Methods (2024)
High-throughput image processing software for the study of nuclear architecture and gene expression
- Adib Keikhosravi
- Faisal Almansour
- Gianluca Pegoraro
Scientific Reports (2024)
BioImage.IO Chatbot: a community-driven AI assistant for integrative computational bioimaging
- Wanlu Lei
- Caterina Fuster-Barceló
- Wei Ouyang
Nature Methods (2024)