The promise and peril of deep learning in microscopy (original) (raw)
- News & Views
- Published: 21 January 2021
MICROSCOPY
Nature Methods volume 18, pages 131–132 (2021)Cite this article
- 6072 Accesses
- 27 Citations
- 95 Altmetric
- Metrics details
Subjects
Ever since van Leeuwenhoek peered into his homemade microscope and revealed a world inhabited by “small animals,” scientists have been pushing the limits of microscopy to see ever finer details in living cells and organisms. Though current methods would be science fiction to van Leeuwenhoek, we’ve entered an era of diminishing returns: camera sensors are 95% efficient, modern lasers can evaporate samples, fluorescent molecules reliably emit thousands of photons, and objectives lenses hit their fundamental physical performance limit over a century ago. Nevertheless, a “resolution revolution” seeded by Lukosz in the 1960s and heralded by Hell, Gustafsson and Betzig in the 2000s introduced “super-resolution” (SR) to the scientific vernacular. In this issue of Nature Methods, Qiao et al.[1](/articles/s41592-020-01035-w#ref-CR1 "Qiao, C. Nat. Methods https://doi.org/10.1038/s41592-020-01048-5
(2021).") continue the revolution by riding the tidal wave of deep learning (DL) — a framework rooted in the 1940s[2](/articles/s41592-020-01035-w#ref-CR2 "McCulloch, W. S. & Pitts, W. Bull. Math. Biophys. 5, 115–133 (1943).") that has only recently enjoyed the computational muscle required by any but the simplest tasks[3](/articles/s41592-020-01035-w#ref-CR3 "Silver, D. et al. Nature 550, 354–359 (2017).") — and present alchemic results: transforming low-resolution, low-contrast, noisy images into super-resolved, high contrast, clean micrographs.Qiao et al.’s achievement is threefold. First, they collect an exceptional training dataset[1](/articles/s41592-020-01035-w#ref-CR1 "Qiao, C. Nat. Methods https://doi.org/10.1038/s41592-020-01048-5
(2021)."), an invaluable public resource for new method development, consisting of matched noisy, low-resolution images and high-quality, super-resolved, structured illumination microscopy (SIM, a variant of SR microscopy) reconstructions. Second, they introduce two DL architectures, termed deep Fourier channel attention networks (DFCAN) and deep Fourier generative adversarial networks (DFGAN), which, as their names imply, learn feature representations in the Fourier domain; and finally, they apply the networks to both the perennial problem of SIM reconstruction from nine low-quality images and the more fantastical concept of single-image SR (SISR)[4](/articles/s41592-020-01035-w#ref-CR4 "Dong, C., Loy, C. C., He, K. & Tang, X. IEEE Trans. Pattern Anal. Mach. Intell. 38, 295–307 (2016)."), in which an SR image is inferred entirely from a single diffraction-limited, or lower resolution, image.This is a preview of subscription content, access via your institution
Relevant articles
Open Access articles citing this article.
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Additional access options:
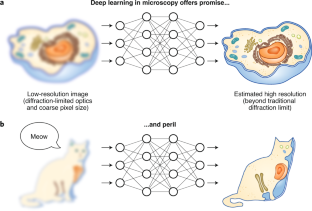
Fig. 1: Do androids dream of electric cells?
References
- Qiao, C. Nat. Methods https://doi.org/10.1038/s41592-020-01048-5 (2021).
- McCulloch, W. S. & Pitts, W. Bull. Math. Biophys. 5, 115–133 (1943).
Article Google Scholar - Silver, D. et al. Nature 550, 354–359 (2017).
Article CAS Google Scholar - Dong, C., Loy, C. C., He, K. & Tang, X. IEEE Trans. Pattern Anal. Mach. Intell. 38, 295–307 (2016).
Article Google Scholar - McElreath, R. Statistical Rethinking: A Bayesian Course with Examples in R and Stan (Taylor and Francis, 2020).
- Citron, D. Deep fakes: a looming challenge for privacy, democracy, and national security. Center for Internet and Society https://cyberlaw.stanford.edu/publications/deep-fakes-looming-challenge-privacy-democracy-and-national-security (2018).
- Wang, H. & Yeung, D.-Y. A survey on Bayesian deep learning. ACM Comput. Surv. 53, 1–37 (2020).
Google Scholar - Eisenstein, M. Nat. Methods 17, 1075–1079 (2020).
Article CAS Google Scholar - Hutson, M. Science https://doi.org/10.1126/science.aau0577 (2018).
- D’Amour, A. et al. Underspecification presents challenges for credibility in modern machine learning. Preprint at arXiv https://arxiv.org/abs/2011.03395 (2020).
Author information
Authors and Affiliations
- 10x Genomics, Pleasanton, CA, USA
David P. Hoffman - DrivenData Inc, Denver, CO, USA
Isaac Slavitt & Casey A. Fitzpatrick
Authors
- David P. Hoffman
You can also search for this author inPubMed Google Scholar - Isaac Slavitt
You can also search for this author inPubMed Google Scholar - Casey A. Fitzpatrick
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toDavid P. Hoffman.
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Hoffman, D.P., Slavitt, I. & Fitzpatrick, C.A. The promise and peril of deep learning in microscopy.Nat Methods 18, 131–132 (2021). https://doi.org/10.1038/s41592-020-01035-w
- Published: 21 January 2021
- Issue Date: February 2021
- DOI: https://doi.org/10.1038/s41592-020-01035-w