Genetic tool development in marine protists: emerging model organisms for experimental cell biology (original) (raw)
- Resource
- Open access
- Published: 06 April 2020
- R. Ellen R. Nisbet ORCID: orcid.org/0000-0003-4487-196X2 na1 nAff54,
- José A. Fernández Robledo3 na1,
- Elena Casacuberta4 na1,
- Lisa Sudek5 na1,
- Andrew E. Allen ORCID: orcid.org/0000-0001-5911-60816,7,
- Manuel Ares Jr ORCID: orcid.org/0000-0002-2552-91688,
- Cristina Aresté4,
- Cecilia Balestreri9,
- Adrian C. Barbrook2,
- Patrick Beardslee10,
- Sara Bender11,
- David S. Booth12,
- François-Yves Bouget13,
- Chris Bowler ORCID: orcid.org/0000-0003-3835-618714,
- Susana A. Breglia15,
- Colin Brownlee9,
- Gertraud Burger ORCID: orcid.org/0000-0002-8679-881216,
- Heriberto Cerutti10,
- Rachele Cesaroni17,
- Miguel A. Chiurillo18,
- Thomas Clemente10,
- Duncan B. Coles3,
- Jackie L. Collier ORCID: orcid.org/0000-0001-8774-571519,
- Elizabeth C. Cooney ORCID: orcid.org/0000-0001-7435-760920,
- Kathryn Coyne21,
- Roberto Docampo ORCID: orcid.org/0000-0003-4229-878418,
- Christopher L. Dupont ORCID: orcid.org/0000-0002-0896-65427,
- Virginia Edgcomb ORCID: orcid.org/0000-0001-6805-381X22,
- Elin Einarsson2,
- Pía A. Elustondo15 nAff55,
- Fernan Federici23,
- Veronica Freire-Beneitez24,25,
- Nastasia J. Freyria ORCID: orcid.org/0000-0002-4972-93833,
- Kodai Fukuda26,
- Paulo A. García27,
- Peter R. Girguis28,
- Fatma Gomaa28,
- Sebastian G. Gornik ORCID: orcid.org/0000-0002-8026-133629,
- Jian Guo5,8,
- Vladimír Hampl30,
- Yutaka Hanawa31,
- Esteban R. Haro-Contreras15,
- Elisabeth Hehenberger20,
- Andrea Highfield9,
- Yoshihisa Hirakawa31,
- Amanda Hopes32,
- Christopher J. Howe ORCID: orcid.org/0000-0002-6975-86402,
- Ian Hu ORCID: orcid.org/0000-0002-2287-031X2,
- Jorge Ibañez23,
- Nicholas A. T. Irwin20,
- Yuu Ishii ORCID: orcid.org/0000-0003-1735-955733,
- Natalia Ewa Janowicz30,
- Adam C. Jones ORCID: orcid.org/0000-0001-7521-186311,
- Ambar Kachale1,
- Konomi Fujimura-Kamada34,
- Binnypreet Kaur1,
- Jonathan Z. Kaye11,
- Eleanna Kazana24,25,
- Patrick J. Keeling20,
- Nicole King12,
- Lawrence A. Klobutcher35,
- Noelia Lander18,
- Imen Lassadi2,
- Zhuhong Li18,
- Senjie Lin ORCID: orcid.org/0000-0001-8831-611135,
- Jean-Claude Lozano13,
- Fulei Luan10,
- Shinichiro Maruyama ORCID: orcid.org/0000-0002-1128-591633,
- Tamara Matute23,
- Cristina Miceli36,
- Jun Minagawa ORCID: orcid.org/0000-0002-3028-320334,37,
- Mark Moosburner6,7,
- Sebastián R. Najle ORCID: orcid.org/0000-0002-0654-91474,38,
- Deepak Nanjappa21,
- Isabel C. Nimmo2,
- Luke Noble39 nAff56,
- Anna M. G. Novák Vanclová30,
- Mariusz Nowacki ORCID: orcid.org/0000-0003-4894-290517,
- Isaac Nuñez23,
- Arnab Pain ORCID: orcid.org/0000-0002-1755-281940,41,
- Angela Piersanti36,
- Sandra Pucciarelli36,
- Jan Pyrih24,30,
- Joshua S. Rest42,
- Mariana Rius19,
- Deborah Robertson43,
- Albane Ruaud23 nAff57,
- Iñaki Ruiz-Trillo4,44,45,
- Monika A. Sigg12,
- Pamela A. Silver ORCID: orcid.org/0000-0002-7856-407146,47,
- Claudio H. Slamovits15,
- G. Jason Smith ORCID: orcid.org/0000-0003-1258-480048,
- Brittany N. Sprecher ORCID: orcid.org/0000-0001-5032-383435,
- Rowena Stern9,
- Estienne C. Swart17 nAff57,
- Anastasios D. Tsaousis ORCID: orcid.org/0000-0002-5424-190524,25,
- Lev Tsypin ORCID: orcid.org/0000-0002-0642-846849,50,
- Aaron Turkewitz ORCID: orcid.org/0000-0003-3531-580649,
- Jernej Turnšek ORCID: orcid.org/0000-0002-9056-35656,7,46,47,
- Matus Valach ORCID: orcid.org/0000-0001-8689-008016,
- Valérie Vergé13,
- Peter von Dassow23,51,
- Tobias von der Haar ORCID: orcid.org/0000-0002-6031-925424,
- Ross F. Waller2,
- Lu Wang52,
- Xiaoxue Wen10,
- Glen Wheeler9,
- April Woods48,
- Huan Zhang35,
- Thomas Mock ORCID: orcid.org/0000-0001-9604-036232,
- Alexandra Z. Worden ORCID: orcid.org/0000-0002-9888-93245,53 &
- …
- Julius Lukeš ORCID: orcid.org/0000-0002-0578-66181
Nature Methods volume 17, pages 481–494 (2020)Cite this article
- 26k Accesses
- 98 Citations
- 311 Altmetric
- Metrics details
Subjects
This article has been updated
Abstract
Diverse microbial ecosystems underpin life in the sea. Among these microbes are many unicellular eukaryotes that span the diversity of the eukaryotic tree of life. However, genetic tractability has been limited to a few species, which do not represent eukaryotic diversity or environmentally relevant taxa. Here, we report on the development of genetic tools in a range of protists primarily from marine environments. We present evidence for foreign DNA delivery and expression in 13 species never before transformed and for advancement of tools for eight other species, as well as potential reasons for why transformation of yet another 17 species tested was not achieved. Our resource in genetic manipulation will provide insights into the ancestral eukaryotic lifeforms, general eukaryote cell biology, protein diversification and the evolution of cellular pathways.
Similar content being viewed by others
Main
The ocean represents the largest continuous planetary ecosystem, hosting an enormous variety of organisms, which include microscopic biota such as unicellular eukaryotes (protists). Despite their small size, protists play key roles in marine biogeochemical cycles and harbor tremendous evolutionary diversity1,2. Notwithstanding their significance for understanding the evolution of life on Earth and their role in marine food webs, as well as driving biogeochemical cycles to maintain habitability, little is known about their cell biology including reproduction, metabolism and signaling3. Most of the biological knowledge available is based on comparison of proteins from cultured species to homologs in genetically tractable model taxa4,5,6,7. A main impediment to understanding the cell biology of these diverse eukaryotes is that protocols for genetic modification are only available for a small number of species8,9 that represent neither the most ecologically relevant protists nor the breadth of eukaryotic diversity.
The development of genetic tools requires reliable information about gene organization and regulation of the emergent model species. Over the last decade, genome4,5,6 and transcriptome sequencing initiatives7 have resulted in nearly 120 million unigenes being identified in protists10, which facilitates the developments of genetic tools used for model species. Insights from these studies enabled the phylogenetically informed approach7 for selecting and developing key marine protists into model systems in the Environmental Model Systems (EMS) Project presented herein. Forty-one research groups took part in the EMS Project, a collaborative effort resulting in the development of genetic tools that significantly expand the number of eukaryotic lineages that can be manipulated, and that encompass multiple ecologically important marine protists.
Here, we summarize detailed methodological achievements and analyze results to provide a synthetic ‘transformation roadmap’ for creating new microeukaryotic model systems. Although the organisms reported here are diverse, the paths to overcome difficulties share similarities, highlighting the importance of building a well-connected community to overcome technical challenges and accelerate the development of genetic tools. The 13 emerging model species presented herein, and the collective set of genetic tools from the overall collaborative project, will not only extend our knowledge of marine cell biology, evolution and functional biodiversity, but also serve as platforms to advance protistan biotechnology.
Results
Overview of taxa in the EMS initiative
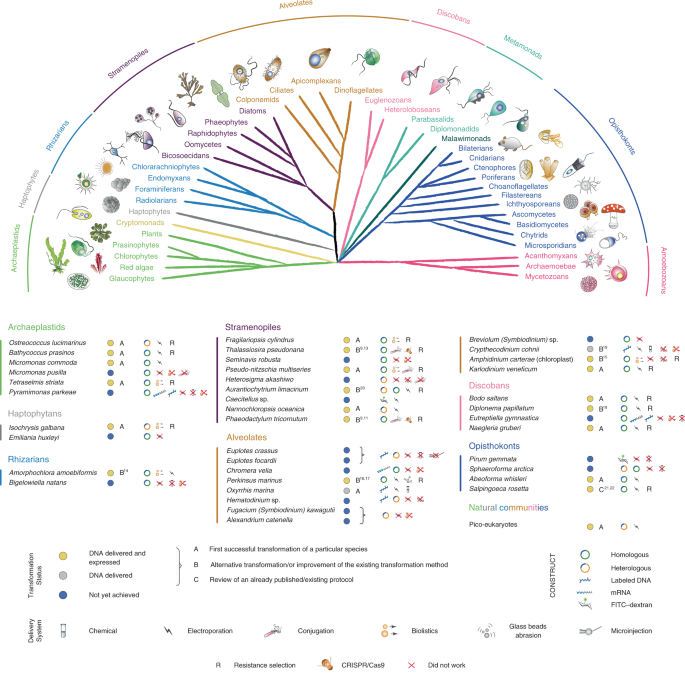
Taxa were selected from multiple eukaryotic supergroups1,7 to maximize the potential of cellular biology and to evaluate the numerous unigenes with unknown functions found in marine protists (Fig. 1). Before the EMS initiative, reproducible transformation of marine protists was limited to only a few species such as Thalassiosira pseudonana, Phaeodactylum tricornutum and Ostreococcus tauri (Supplementary Table 1). The EMS initiative included 39 species, specifically, 6 archaeplastids, 2 haptophytes, 2 rhizarians, 9 stramenopiles, 12 alveolates, 4 discobans and 4 opisthokonts (Fig. 1). Most of these taxa were isolated from coastal habitats, the focus area of several culture collections7. More than 50% of the selected species are considered photoautotrophs, with another 35% divided between heterotrophic osmotrophs and phagotrophs, the remainder being predatory mixotrophs. Almost 20% of the chosen species are symbionts and/or parasites of marine plants or animals, 5% are associated with detritus and several are responsible for harmful algal blooms (Supplementary Table 2).
Fig. 1: Phylogenetic relationships and transformation status of marine protists.
A schematic view of the eukaryotic tree of life with effigies of main representatives. Color-coordinated species we have attempted to genetically modify are listed below. Current transformability status is schematized in circles indicating: DNA delivered and shown to be expressed (yellow, for details see text and Table 1); DNA delivered, but no expression seen (gray) and no successful transformation achieved despite efforts (blue). The details of transformation of species that belong to ‘DNA delivered’ and ‘Not achieved yet’ categories are described in Supplementary Table 5. mRNA, messenger RNA; FITC–dextran, fluorescein isothiocyanate (FITC)-conjugated dextran.
While some transformation systems for protists have been developed in the past8,9,11, the challenge for this initiative was to develop genetic tools for species that not only require different cultivation conditions but are also phenotypically diverse. It should be noted that not all main lineages were explored. For example, amoebozoans did not feature in this aquatic-focused initiative, in part because they tend to be most important in soils, at least based on current knowledge, and manipulation systems exist for members of this eukaryotic supergroup, such as Dictyostelium discoideum12. The overall EMS initiative outcomes are summarized in Fig. 1 and Table 1. We provide detailed protocols for 13 taxa, for which no transformation systems have been previously reported (category A) and eight taxa, for which existing protocols9,11,13,14,15,16,17,18,19,20,21 were advanced (category B; Figs. 2, 3 and 4, Table 1, Supplementary Tables 1–5 and Methods). We also review an already published EMS transformation protocol22 in one species (category C), and we discuss unsuccessful transformation attempts for 17 additional taxa (Fig. 1 and Methods). Finally, we synthesize our findings in a roadmap for the development of transformation systems in protists (Fig. 5).
Table 1 Parameters used for successful transformation as shown in Figs. 2, 3 and 4
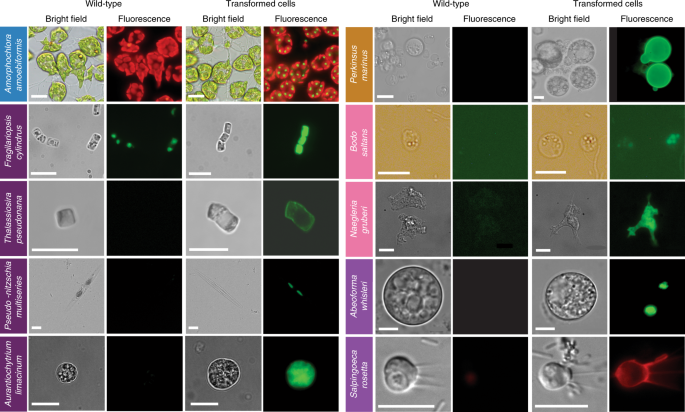
Fig. 2: Epifluorescence micrographs of transformed marine protists.
Representative images of transformants and wild-type cell lines of ten selected protist species. Colored boxes behind species names refer to phylogenetic supergroup assignments given in Fig. 1. Representative data of at least two independent experiments are shown. The fluorescent images show the expression of individual fluorescent marker genes introduced via transformation for all organisms shown, except in the case of A. amoebiformis. For this, red depicts the natural autofluorescence of photosynthetic pigments in the cell, while the additional green spheres in the transformant fluorescence panel shows introduced GFP fluorescence (see Supplementary Fig. 15c for a trace of these different regions in the cell). Scale bars are as follows: 10 µm for A. amoebiformis, T. pseudonana, A. limacinum, B. saltans, N. gruberi, A. whisleri and S. rosetta; 15 µm for P. marinus; 20 µm for F. cylindrus and 100 µm for P. multiseries.
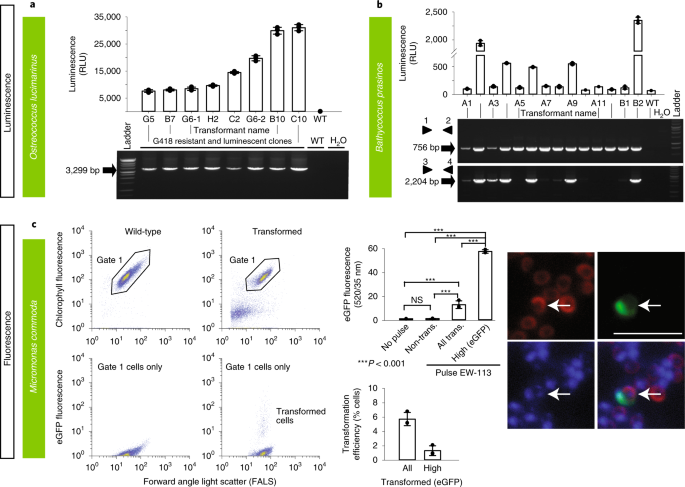
Fig. 3: Various methods were used to demonstrate successful transformation in different archaeplastid species: luminescence and fluorescence.
a–c, Luminescence (a,b) and fluorescence (by FACS and epifluorescence microscopy) (c) were used to verify expression of introduced constructs in three archaeplastids: O. lucimarinus (a), B. prasinos (b) and M. commoda (c). For the latter, red in the image depicts the natural autofluorescence of photosynthetic pigments in the cell, while green shows introduced eGFP fluorescence and blue shows the DAPI-stained nucleus; the overlay shows colocalization of eGFP and nucleus signals. See Supplementary Fig. 15d for a trace of these different regions in the cell. NS, not significant; trans., transformed. Representative data of at least two independent experiments are shown. For a detailed figure description see Supplementary Notes 2.
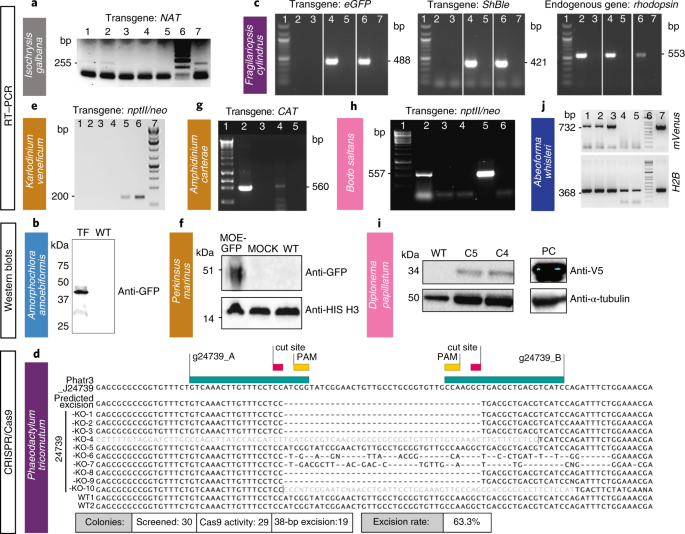
Fig. 4: Various methods were used to demonstrate successful transformation in different species: RT–PCR, western blot and sequencing.
a–j, Western blot, RT–PCR or sequencing (in case of Cas9-induced excision by CRISPR) were used to verify expression of introduced constructs in one haptophyte: I. galbana (a), one rhizarian—A. amoebiformis (b), two stramenopiles—F. cylindrus (c) and P. tricornutum (d), three alveolates—K. veneficum (e), P. marinus (f) and A. carterae (g), two discobans—B. saltans (h) and D. papillatum (i) and one opisthokont—A. whisleri (j). Note that nptII/neo is used synonymously with amino 3′-glycosyl phosphotransferase gene (aph(3′)) conferring resistance to kanamycin and neomycin. Representative data of at least two independent experiments are shown. For a detailed figure description see Supplementary Notes 2.
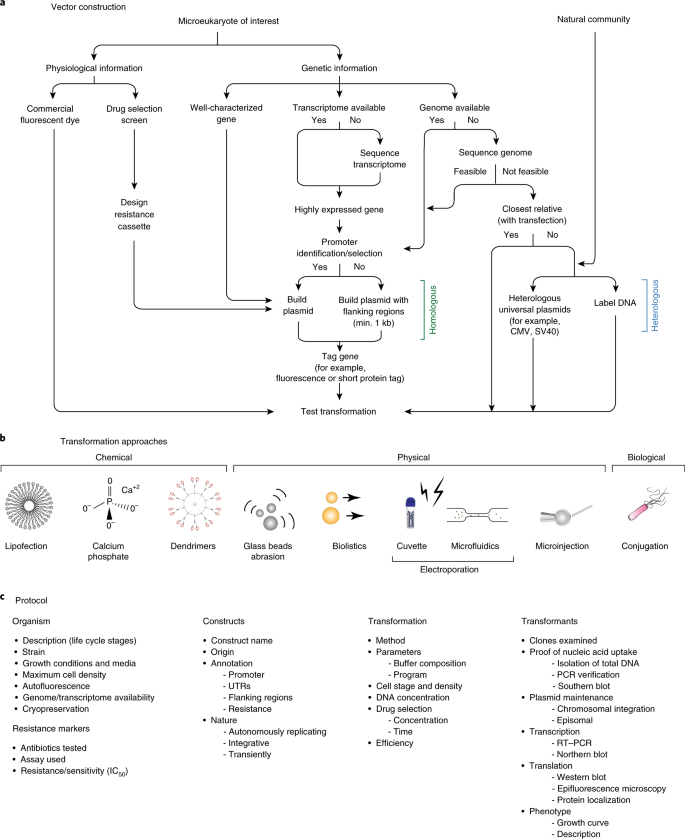
Fig. 5: ‘Transformation roadmap’ for the creation of genetically tractable protists.
a, Vector design and construction for microeukaryotes of interest and a natural community. b, Transformation approaches. Different symbols represent methods (for example chemical, physical or biological) for introducing DNA/RNA/protein into a living cell. c, Protocol. Key methodological steps for successful transformation are listed in an abbreviated form (for particular examples, see Table 1 and text).
Archaeplastids
Prasinophytes are important marine green algae distributed from polar to tropical regions23. They form a sister group to chlorophyte algae, and together, these two groups branch adjacent to land plants, collectively comprising the Viridiplantae, which are part of the Archaeplastida1,23 (Fig. 1). Genome sequences are available for the picoprasinophytes (<3 µm cell diameter) tested herein, specifically, Micromonas commoda, M. pusilla, Ostreococcus lucimarinus and Bathycoccus prasinos. As part of the EMS initiative, we report on genetic tools for Bathycoccus, a scaled, nonmotile genus, and Micromonas, a motile, naked genus with larger genomes than Bathycoccus and Ostreococcus22. We also report on genetic tools for Tetraselmis striata and O. lucimarinus. The latter was transformed based on an adapted homologous recombination system for O. tauri24,25.
O. lucimarinus (RCC802) and B. prasinos (RCC4222) were transformed using protocols adapted from O. tauri24,25. Briefly, using electroporation for transfer of exogenous genes, O. lucimarinus was transformed using a DNA fragment encoding the O. tauri high-affinity phosphate transporter (HAPT) gene fused to a luciferase gene and a kanamycin selection marker (Table 1 and Supplementary Table 3), which resulted in transient luciferase expression 24 h after electroporation (Table 1 and Fig. 3a). After 2 weeks of growth in low-melting agarose plates containing G418 (1 mg ml−1), 480 colonies were obtained, picked and grown in artificial seawater with the antibiotic neomycin. Of these, 76 displayed luminescence ≥2.5-fold above background (80 relative luminescence units (RLU)), with widely variable levels (200–31,020 RLU), likely reflecting either variations in the site of integration and/or the number of integrated genes (Fig. 3a, Supplementary Fig. 1 and Methods).
The O. tauri construct did not work in B. prasinos, while the use of the B. prasinos histone H4 and HAPT sequences in an otherwise identical construct and conditions was successful. Although luciferase expression was not detected 24 h after electroporation, 48 G418-resistant colonies were obtained 2 weeks later, 20 being luminescent when grown in liquid medium. Analysis of 14 resistant transformants revealed that the luciferase sequence was integrated into the genome of five luminescent clones, and one nonluminescent clone (Fig. 3b and Methods), suggesting that the chromatin context at integration sites in the latter was not favorable to luciferase expression.
Although transformation methods successful for Bathycoccus and Ostreococcus failed in Micromonas, Lonza nucleofection was successful with M. commoda (CCMP2709) (Table 1 and Fig. 3c) using two different codon-optimized plasmids, one encoding the luciferase gene (NanoLuc, Promega) flanked by an exogenous promoter and terminator sequence from the 5′ and 3′ untranslated regions (UTRs) of histone H3 in Micromonas polaris (CCMP2099), and the other encoding an enhanced green fluorescent protein (eGFP) gene flanked by endogenous promoter and terminator sequences from ribosomal protein S9 (Supplementary Table 5). Sensitivities to antibiotics were established (Supplementary Table 3). Constructs did not include a selectable marker, as we aimed to introduce and express foreign DNA while developing conditions suitable for transfection that supported robust growth in this cell wall-lacking protist (Table 1). Transformants revealed a significantly higher level of eGFP fluorescence than wild-type cells, with 1.3% of the population showing fluorescence per cell 45-fold higher than both the nontransformed portion of the culture and the wild-type cells (Fig. 3c and Methods). Additionally, the RLU was 1,500-fold higher than controls when using the luciferase-bearing construct, such that multiple experiments with both plasmids confirmed expression of exogenous genes in M. commoda.
T. striata (KAS-836) was transformed using microprojectile bombardment (Supplementary Fig. 2a). Two selectable marker genes were tested, consisting of a putative promoter and 5′ UTR sequences from the T. striata actin gene and either the coding sequences of the Streptoalloteichus hindustanus bleomycin gene (conferring resistance to zeocin) or the Streptomyces hygroscopicus bar gene (conferring resistance to glufosinate) (Table 1, Supplementary Fig. 2a and Methods). The terminator sequence was obtained from the T. striata glyceraldehyde-3-phosphate dehydrogenase gene. Linearized plasmids were coated on gold particles and introduced into T. striata cells by using the PDS-1000/He Particle Delivery System (Bio-Rad). Transformants were successfully selected on half-strength f/2 at 50% salinity agar plates containing either 150 μg ml−1 zeocin or 150 μg ml−1 glufosinate.
Haptophytes (incertae sedis)
Haptophytes are a group of photosynthetic protists that are abundant in marine environments and include the principal calcifying lineage, the coccolithophores. Genome sequences are available for Emiliania huxleyi6 and Chrysochromulina tobin26, and there is one report of nuclear transformation of a calcifying coccolithophore species27 but transformation of E. huxleyi, the most prominent coccolithophore, has not been achieved yet27. Here, as part of the EMS initiative, a stable nuclear transformation system was developed for Isochrysis galbana, a species that lacks coccoliths, but represents an important feedstock for shellfish aquaculture28.
I. galbana (CCMP1323) was transformed by biolistic bombardment with the pIgNAT vector, which contains nourseothricin (NTC) _N_-acetyltransferase (NAT), (for nourseothricin resistance) driven by the promoter and terminator of Hsp70 from E. huxleyi (CCMP1516). Twenty-four hours after bombardment, cells were transferred to liquid f/2 medium at 50% salinity containing 80 µg ml−1 NTC and left to grow for 2–3 weeks to select for transformants (Table 1). The presence of NAT in NTC-resistant cells was verified by PCR and PCR with reverse transcription (RT–PCR) (Fig. 4a, Supplementary Fig. 2b and Methods) and the sequence was verified. To confirm NTC resistance was a stable phenotype, cells were subcultured every 2–4 weeks at progressively higher NTC concentrations (up to 150 µg ml−1) in the above-mentioned media. Cells remained resistant to NTC for approximately 6 months, as confirmed by PCR screening to identify the presence of the NAT gene.
Rhizarians
Rhizarians include diverse nonphotosynthetic protists, as well as the photosynthetic chlorarachniophytes that acquired a plastid via secondary endosymbiosis of a green alga4. Uniquely, they represent an intermediate stage of the endosymbiotic process, since their plastids still harbor a relict nucleus (nucleomorph). Here, we report on an advanced transformation protocol for the chlorarachniophyte Amorphochlora (Lotharella) amoebiformis for which low-efficiency transient transformation has previously been achieved using particle bombardment14.
A. amoebiformis (CCMP2058) cells were resuspended in 100 µl of Gene Pulse Electroporation Buffer (Bio-Rad) with 20–50 µg of the reporter plasmid encoding eGFP-RubisCO fusion protein under the control of the native rbcS1 promoter and subjected to electroporation (Table 1). Cells were immediately transferred to fresh ESM medium and incubated for 24 h. Transformation efficiency was estimated by the fraction of cells expressing eGFP, resulting in 0.03–0.1% efficiency, as enumerated by microscopy, showing an efficiency up to 1,000-fold higher than in the previous study14 (Table 1). Stable transformants were generated by manual isolation using a micropipette, and a transformed line has maintained eGFP fluorescence for at least 10 months without antibiotic selection (Figs. 2 and 4b and Methods).
Stramenopiles
Stramenopiles are a diverse lineage harboring important photoautotrophic, mixotrophic (combining photosynthetic and phagotrophic nutrition) and heterotrophic taxa. As the most studied class in this lineage, diatoms (Bacillariophyceae) were early targets for the development of reverse genetics tool11,29. Diatoms are estimated to contribute approximately 20% of annual carbon fixation30 and, like several other algal lineages, are used in bioengineering applications and biofuels31. Although other cold-adapted eukaryotes have, to our knowledge, yet to be transformed, here we present a protocol for the Antarctic diatom Fragilariopsis cylindrus32. A transformation protocol has also been developed herein for Pseudo-nitzschia multiseries, a toxin-producing diatom33. Here we also present work for nondiatom stramenopiles, including a transformation protocol for the eustigmatophyte Nannochloropsis oceanica, and an alternative protocol for the labyrinthulomycete Aurantiochytrium limacinum20, both of which are used for biotechnological applications. Furthermore, we report on advances for CRISPR/Cas-driven gene knockouts in Phaeodactylum tricornutum8,13 and a more efficient bacterial conjugation system for Thalassiosira pseudonana13.
Microparticle bombardment was used on F. cylindrus (CCMP1102) that was grown, processed and maintained at 4 °C in 24 h light. Exponential phase cells were harvested onto a 1.2 µm membrane filter that was then placed on an 1.5% agar Aquil plate for bombardment with beads coated with a plasmid containing zeocin resistance and eGFP, both controlled by an endogenous fucoxanthin chlorophyll a/c binding protein (FCP) promoter and terminator (Table 1, Supplementary Table 3 and Methods)34. Transformation was performed using 0.7 µm tungsten particles and the biolistic particle delivery system PDS-1000/He (Bio-Rad). Rupture disks for 1,350 and 1,550 pounds per square inch (psi) gave the highest colony numbers with efficiencies of 20.7 colony forming units (c.f.u.) per 108 cells and 30 c.f.u. per 108 cells, respectively. Following bombardment, the filter was turned upside down and left to recover for 24 h on the plate, then cells were rinsed from the plate/filter and spread across five 0.8% agar Aquil plates with 100 µg ml−1 zeocin. Colonies appeared 3–5 weeks later. PCR on genomic DNA showed that 100 and 60% of colonies screened positive for the bleomycin gene (ShBle) for zeocin resistance and the gene encoding eGFP, respectively. As confirmed by fluorescence-activated cell sorting (FACS) and microscopy, eGFP was localized to the cytosol and was distinguishable from plastid autofluorescence (Fig. 2). Additional confirmation by PCR and RT–PCR (Fig. 4c) revealed that the ShBle and eGFP genes were present in the genomes of transformants after multiple transfers (>10) 2 years later, indicating long-term stability.
Bacterial conjugation methods were improved in T. pseudonana (CCMP1335) using the silaffin precursor TpSil3p (Table 1 and Methods) as the target gene. TpSil3p was fused to eGFP flanked by an FCP promoter and terminator, cloned into a pTpPuc3 episomal backbone and transformed into mobilization plasmid-containing EPI300 E. coli cells (Lucigen). The donor cells were grown in super optimal broth with catabolite repression (SOC) medium at 37 °C until OD600 of 0.3–0.4, centrifuged and resuspended in 267 μl SOC medium. Next, 200 μl of donor cells were mixed with T. pseudonana cells, cocultured on predried 1% agar plates, dark incubated at 30 °C for 90 min, then at 18 °C in constant light for 4 h, followed by selection in 0.25% agar plates containing 100 µg ml−1 NTC. Colonies were observed after 2 weeks, inoculated into 300 μl L1 medium and supplemented with 200 µg ml−1 NTC to reduce the number of false positives. Positive transformants were identified by colony PCR screening (Supplementary Fig. 3) and epifluorescence microscopy (Fig. 2).
The diatom P. multiseries (15093C) and other members of this genus form buoyant linear chains with overlapping cell tips during active growth, and were unconducive to punctate colony formation on agar, where their growth is generally poor. To address this challenge, a low-gelation-temperature agarose seawater medium (LGTA) was developed to facilitate growth, antibiotic selection and cell recovery. P. multiseries exhibited growth inhibition at relatively low concentrations under NTC, formaldehyde and zeocin (Supplementary Table 3). Biolistic transformation of two other P. species had been demonstrated at low efficiency35. To complement this approach and explore potentially higher efficiency methods for transformation with diatom episomal plasmids, we modified the existing conjugation-based method13. The published conjugation protocol was modified to enhance P. multiseries postconjugation viability by reducing SOC content. An episomal version of the Pm_actP_egfp_actT expression cassette was transfected into E. coli EPI300+pTAMOB and used for conjugation (Table 1 and Methods). After 48 h in L1 medium, cells were plated in LGTA and eGFP-positive cells were observed 7 d later (Fig. 2). PCR revealed the presence of plasmids in all eGFP-positive colonies (Supplementary Fig. 4). Similarly, conjugation with the episome pPtPUC3 (bleomycin selection marker)-containing bacterial donors was followed under zeocin selection (200 μg ml−1). After 7 d, only viable cells (based on bright chlorophyll fluorescence) contained the episome, as confirmed by PCR. Propagation of transformants after the first medium transfer (under selection) has so far been unsuccessful.
Stable transformation of A. limacinum (ATCC MYA-1381) was achieved by knock-in of a resistance cassette composed of ShBle driven by 1.3 kb promoter and 1.0 kb terminator regions of the endogenous glyceraldehyde-3-phosphate dehydrogenase gene carried in a pUC19-based plasmid (18GZG) along with the native 18S ribosomal RNA gene, and by knock-in of a similar construct containing a eGFP:ShBle fusion (Supplementary Fig. 5). Approximately 1 × 108 cells were electroporated, adapting the electroporation protocol used for Schizochytrium36. The highest transformation efficiency was achieved using 1 µg of linearized 18GZG plasmid with two pulses, resulting in a time constant of ~5 ms (Table 1 and Methods). Expression of the fusion protein was confirmed by both the zeocin-resistance phenotype and the detection of eGFP (Fig. 2). Six 18GZG transformants derived from uncut and linearized plasmids were examined in detail. All maintained antibiotic resistance throughout 13 serial transfers, first in selective, then subsequently in nonselective media and then again in selective medium. Integration of the plasmid into the genome was confirmed by PCR as well as by Southern blots using a digoxigenin-labeled ShBle gene probe, showing that four transformants had integrations by single homologous recombination, while in two transformants additional copies of the antibiotic resistance cassette were integrated by nonhomologous recombination elsewhere in the genome (Supplementary Fig. 5).
Electroporation of N. oceanica (CCMP1779) was optimized based on observation of cells treated with fluorescein-conjugated 2,000 kDa dextran and subsequent survival (Table 1 and Methods). A sorbitol concentration of 800 mM and electroporation at between 5 and 9 kV cm−1 resulted in highest cell recovery. These conditions were used during introduction of plasmids containing the gene for the blue fluorescent reporter mTagBFP2 under the control of cytomegalovirus (CMV), the cauliflower mosaic virus 35S, or the VCP1 promoter previously described from Nannochloropsis sp.37. Transient expression of blue fluorescence (compared to cells electroporated simultaneously under the same conditions without plasmid) appeared within 2 h, lasted for at least 24 h and disappeared by 48 h in subsets of cells electroporated with mTagBFP2 under the control of CMV (Supplementary Fig. 6). The transient transformation was more effective when a linearized plasmid was used compared to a circular plasmid (Table 1). VCP1 did not induce blue fluorescence with a circular plasmid, while 35S gave inconsistent results with either circularized or linearized plasmids.
For P. tricornutum (CCAP1055/1), we adapted the CRISPR/Cas9 system8 for multiplexed targeted mutagenesis. Bacterial conjugation13 was used to deliver an episome that contained a Cas9 cassette and two single-guide RNA (sgRNA) expression cassettes designed to excise a 38 basepair-long domain from the coding region of a nuclear-encoded, chloroplastic glutamate synthase (Phatr3_J24739) and introduce an in-frame stop codon after strand ligation (Table 1 and Methods). The GoldenGate assembly was used to clone two expression cassettes carrying sgRNAs into a P. tricornutum episome that contained a Cas9–2A-ShBle expression cassette and the centromeric region CenArsHis (Supplementary Fig. 7). After their addition to a P. tricornutum culture, plates were incubated in a growth chamber under standard growth conditions for 2 d and transformed P. tricornutum colonies began to appear after 2 weeks. Only colonies maintaining Cas9–2A-ShBle sequence on the delivered episome were able to grow on selection plates because Cas9 and ShBle were transcriptionally fused by the 2A peptide38 (Supplementary Fig. 7). Gel electrophoresis migration and sequencing of the genomic target loci confirmed the 38 bp-long excision and premature stop codon (Fig. 4d).
Alveolates
This species-rich and diverse group comprises ciliates, apicomplexans and dinoflagellates (Fig. 1). As a link between apicomplexan parasites and dinoflagellate algae, perkinsids are key for understanding the evolution of parasitism, and also have potential biomedical applications17. Techniques currently exist for transformation of only a small number of ciliates, perkinsids and apicomplexans39. Here, we present a transformation protocol for Karlodinium veneficum (CCMP1975), a phagotrophic mixotroph that produces fish-killing karlotoxins40. Experiments were also performed on Oxyrrhis marina (CCMP 1788/CCMP 1795), a basal-branching phagotroph that lacks photosynthetic plastids and Crypthecodinium cohnii (CCMP 316), a heterotroph used in food supplements. For both of these taxa, evidence of DNA delivery was achieved (Table 1, Supplementary Results, Supplementary Fig. 15 and Methods), a goal recently achieved for C. cohnii using electroporation19. Additionally, we report on improved transformation systems for Perkinsus marinus (PRA240) and Amphidinium carterae (CCMP1314) chloroplast, published recently as part of the EMS initiative15.
K. veneficum (CCMP1975) was transformed based on electroporation and cloning the selectable marker gene aminoglycoside 3′-phosphotransferase (nptII/neo; note that nptII/neo is used synonymously with amino 3′-glycosyl phosphotransferase gene conferring resistance to kanamycin, neomycin, paromomycin, ribostamycin, butirosin and gentamicin B) into the backbone of the dinoflagellate-specific expression vector DinoIII-neo41, which confers resistance to neomycin and kanamycin (Table 1). In brief, DinoIII-neo was linearized and electroporated using the Nucleofector optimization pulse codes, buffer SF/Solution I (Lonza), and 2 μg μl−1 of linearized DinoIII-neo. Electroporated cells were selected under 150 μg ml−1 kanamycin 3 d postelectroporation. Fresh seawater with kanamycin was added every 2 weeks to the cultures and new subcultures were inoculated monthly. After 3 months, DNA and RNA were isolated from the resistant cultures as previously reported42 and cDNA was synthesized using random hexamers. Out of 16 transformations, two cell lines (CA-137, DS-138) showed stable growth under kanamycin selection. CA-137 developed dense cultures after 3 months, and the resistance gene was detected in both DNA and RNA by nested PCR and RT–PCR, respectively (Fig. 4e, Supplementary Fig. 8 and Methods).
We improved the transformation protocol16,17 of P. marinus, a pathogen of marine mollusks, fish and amphibians43 (Supplementary Table 5). We coexpressed two genes and efficiently selected transient and stable transformants using FACS (Table 1, Figs. 2 and 4f, Supplementary Fig. 9 and Methods). In addition, we established the integration profile of ectopic DNA once introduced into the P. marinus genome. We did not see evidence of integration through homologous recombination and observed a propensity for plasmid fragmentation and integration within transposable elements sites. An optimized alternative protocol for transformation using glass bead abrasion was also developed. Two versions of the previously published Moe gene promoter16 were tested. Whereas the 1.0 kb promoter version induced expression after 2 or 3 d, the truncated version (0.5 kb) took 7 d for expression to be detected. Resistance genes to zeocin, blasticidin and puromycin have all been shown to confer resistance to transformed P. marinus; however, selection regimes are still relatively slow and inefficient, indicating further room for improvement17.
We also report a vector for the transformation of the A. carterae chloroplast, a photosynthetic dinoflagellate. A. carterae, like other dinoflagellates with a peridinin-containing chloroplast, contains a fragmented chloroplast genome made up of multiple plasmid-like minicircles40. The previous transformation protocols made use of this to introduce two vectors based on the psbA minicircle15. Here, we show that other minicircles are also suitable for use as vectors. We created an artificial minicircle, using the atpB minicircle as a backbone, but replacing the atpB gene with a codon-optimized chloramphenicol acetyltransferase (Table 1 and Methods). This circular vector was introduced by biolistics to A. carterae (Supplementary Fig. 10a). Following selection with chloramphenicol, we were able to detect transcription of the chloramphenicol acetyltransferase gene via RT–PCR (Fig. 4g). This result suggests that all 20 or so minicircles in the dinoflagellate chloroplast genome would be suitable for use as artificial minicircles, thus providing a large pool of potential vectors.
Discobans
This diverse group, recently split into Discoba and Metamonada44, includes heterotrophs, photoautotrophs and predatory mixotrophs, as well as parasites. Discobans include parasitic kinetoplastids with clinical significance, such as Trypanosoma brucei, T. cruzi and Leishmania spp., for which efficient transformation protocols are available45. However, such protocols are missing for aquatic species. Here, we describe available transformation protocols for the kinetoplastid Bodo saltans and the heterolobosean Naegleria gruberi. The former was isolated from a lake, but identical 18S rRNA gene sequences have been reported from marine environments46. The latter is a freshwater protist that represents a model organism for closely related marine heterolobosean amoebas. Furthermore, we provide advanced methods that build on previous EMS results18 for the diplonemid Diplonema papillatum.
B. saltans (ATCC 30904) was transformed with a plasmid containing a cassette designed to fuse an endogenous EF1-α gene with eGFP for C-terminal tagging. This cassette includes downstream of eGFP, a B. saltans tubulin intergenic region followed by the selectable marker nptII/neo gene, conferring resistance to neomycin. EF1-α genes exist in tandem repeats. The homologous regions that flank the cassette were chosen as targets for inducing homology-directed repair; however, they target only one copy of the gene. As transcription in B. saltans is polycistronic46, insertion of the tubulin intergenic region into the plasmid is essential for polyadenylation of the EF1-α/GFP fusion and _trans_-splicing of the nptII/neo gene (Supplementary Table 5). Selection of transfected cells began with 2 µg ml−1 of neomycin added 24 h after electroporation, and this concentration was gradually increased over 2 weeks to 5 µg ml−1 (Table 1 and Methods). Cells were washed and subcultured into fresh selection medium every 4 d, and neomycin-resistant cells emerged 7–9 d postelectroporation. The eGFP signal was detected 2 d postelectroporation, albeit with low intensity. This may be due to the inefficient translation of eGFP since it has not been codon-optimized for B. saltans (Fig. 2). Genotyping analysis 9 months posttransfection confirmed the presence of the nptII/neo gene and at least partial plasmid sequence (Fig. 4h and Supplementary Fig. 10b). However, plasmid integration into the B. saltans genome through homologous recombination is still unconfirmed. This suggests either off-target plasmid integration or episomal maintenance.
For N. gruberi (ATCC 30224) two plasmids were designed. The first one carried the hygromycin B resistance gene (hph) with an actin promoter and terminator, along with an HA-tagged eGFP driven by the ubiquitin promoter and terminator. The second plasmid carried the nptII/neo gene instead. For each individual circular plasmid, 4 μg was electroporated (Table 1 and Methods). About 48 h after electroporation, dead cells were removed from the suspension and viable cells were washed with PBS. Afterward, 300 μg ml−1 of hygromycin B or 700 μg ml−1 of neomycin was added to the fresh media. One to 4 weeks later, resistant clones were recovered and expression of eGFP and/or hygromycin was confirmed by western blotting (Supplementary Fig. 11). Expression of eGFP was observed by epifluorescence microscopy (Fig. 2 and Supplementary Fig. 11) with ~80% of transformants maintaining hygromycin B or neomycin resistance in addition to expressing eGFP.
D. papillatum (ATCC 50162) was transformed by electroporation using 3 μg of a _SwaI_-linearized fragment (cut from p57-V5+NeoR plasmid) containing the V5-tagged nptII/neo gene flanked by partial regulatory sequences derived from the hexokinase gene of the kinetoplastid Blastocrithidia (strain p57) (Table 1 and Methods) using a published protocol18. About 18 h after electroporation, 75 μg ml−1 G418 was added to the medium and after 2 weeks, seven neomycin-resistant clones were recovered. Transcription of nptII/neo was verified in four clones by RT–PCR (Supplementary Fig. 12) and the expression of the tagged nptII/neo protein was confirmed in two clones by western blotting using the α-V5 antibody (Fig. 4i).
Opisthokonts
The opisthokont clade Holozoa includes animals and their closest unicellular relatives choanoflagellates, filastereans, ichthyosporeans and corallochytreans. The establishment of genetic tools in nonmetazoan holozoans promises to help illuminate the cellular and genetic foundations of animal multicellularity47. Genomic and transcriptomic data are available for multiple representatives characterized by diverse cell morphologies, some of which can even form multicellular structures46. Until recently, only transient transformations had been achieved for some opistokonts such as the filasterean Capsaspora owczarzaki48, the ichthyosporean Creolimax fragrantissima49 and the choanoflagellate Salpingoeca rosetta21. Through the EMS initiative, we report on evidence for transient transformation of the ichthyosporean Abeoforma whisleri, isolated from the digestive tract of mussels, and review a recently published stable transformation protocol for S. rosetta achieved by using the selectable puromycin _N_-acetyl-transferase gene (Fig. 2)22.
All A. whisleri life stages are highly sensitive to a variety of methods for transformation. However, we developed a 4D-nucleofection-based protocol using 16-well strips, wherein PBS-washed cells were resuspended in 20 μl of buffer P3 (Lonza) containing 40 μg of carrier plasmid (empty pUC19) and 1–5 μg of the reporter plasmid (A. whisleri H2B fused to mVenus fluorescent protein, mVFP) (Table 1 and Methods), and subjected to code EN-138 (Lonza). Immediately after the pulse, cells were recovered by adding 80 μl of marine broth (Gibco) before plating in 12-well culture plates previously filled with 1 ml marine broth. After 24 h, ~1% of the culture was transformed based on the fraction of cells expressing mVFP in the nucleus (Figs. 2 and 4j).
Microbial eukaryotes in natural planktonic communities
Model organisms are typically selected based on criteria such as relative ease of isolation and asexual cultivation in the laboratory; however, these attributes may not correlate with the capacity for uptake and expression of the exogenous DNA. We explored whether natural marine planktonic pico- and nanoeukaryote communities would take up DNA in a culture-independent setting. Microbial plankton from natural seawater was concentrated and electroporated with plasmids containing mTagBFP2 under the control of CMV or 35S promoters (Supplementary Results and Methods). In most trials, blue fluorescent cells were rare if detected at all (compared to control samples). However, in one natural community tested, a photosynthetic picoeukaryote population exhibited up to 50% of cells with transient expression of blue fluorescence when the CMV promoter was used (Supplementary Fig. 13). This suggests it might be possible to selectively culture eukaryotic microorganisms based on capacity to express exogenous DNA.
Discussion
The collaborative effort by the EMS initiative facilitated identification and optimization of the steps required to create new protist model systems, which culminated in the synthetic transformation roadmap (Fig. 5). Our genetic manipulation systems for aquatic (largely marine) protists will enable deeper insights into their cell biology, with potentially valuable outcomes for aquatic sciences, evolutionary studies, nanotechnology, biotechnology, medicine and pharmacology. Successes and failures with selectable markers, transformation conditions and reporters were qualitatively compared across species (Supplementary Tables 3 and 4, Table 1, Figs. 2–4 and Methods).
For some of the selected species, the first step was to identify cultivation conditions for robust growth in the laboratory to either generate high cell densities or large culture volumes for obtaining sufficient biomass required for a variety of molecular biology experiments. Unlike established microbial model species, cultivation of marine protists can be challenging, especially under axenic conditions or for predatory taxa that require cocultivation with their prey. Nevertheless, 13 out of 35 species were rendered axenic before the development of transformation protocols. For the remaining species, we were unable to remove bacteria and therefore had to make sure that transformation signals were coming from the targeted protist rather than contaminants (Supplementary Table 2). Subsequent steps included the identification of suitable antibiotics and their corresponding selectable markers (Table 1 and Supplementary Table 3), conditions for introducing exogenous DNA (Table 1 and Supplementary Table 4) and selection of promoter and terminator sequences for designing transformation vectors (Table 1, Methods, Supplementary Table 5 and Supplementary Notes 1).
As exemplified in the model systems provided herein (Table 1 and Figs. 2–4), a variety of methods were used to test whether exogenous DNA was integrated into the genome or maintained as a plasmid, and whether the introduced genes were expressed. Approaches to show the former included inverse PCR, Southern blotting and whole genome sequencing, whereas approaches to demonstrate the latter included various combinations of PCR, RT–PCR, western blotting, epifluorescence microscopy, FACS, antibody-based methods and/or growth assays in the presence of antibiotics to confirm transcription and translation of introduced selection and reporter genes (for example, eGFP, YFP, mCherry). For fluorescent markers, it was first ensured that the wild-type, or manipulated controls cells, had no signals conflicting with the marker (Figs. 2 and 3c), an important step because photosynthetic protists contain chlorophyll and other autofluorescent pigments. Overall transformation outcomes for each species were parsed into three groups according to the level of success or lack thereof (A, first transformation protocol for a given species; B, advanced protocol based on previous work and C, published protocol based on the EMS initiative) and are discussed according to their phylogenetic position (Fig. 1).
Our studies did not result in a universally applicable protocol because transformability and a range of other key conditions varied greatly across taxa and approaches, such as intrinsic features of the genome and differences in cellular structure and morphology. In general, electroporation proved to be the most common method for introducing exogenous DNA stably into cells. This approach was used for naked cells and protoplasts, yet frequently also worked, albeit with lower efficiency, on cells protected by cell walls. Linearized plasmids were most effective for delivery, and 5′ and 3′ UTR-containing promotors of highly expressed endogenous genes provided the strongest expression of selective reporters and markers. If successful, teams usually continued with fluorescence-based methods. Furthermore, large amounts of carrier DNA usually facilitated successful initial transformations (for example, M. commoda, A. whisleri) or improved existing protocols (S. rosetta21). We also provide the contact details of all coauthors who are assigned to particular species (Supplementary Table 6).
Some lineages were difficult to transform, especially dinoflagellates and coccolithophores. Here, even if DNA appeared to be delivered (Supplementary Table 5), expression of the transformed genes could not be confirmed. Examples include the dinoflagellates C. cohnii, Symbiodinium microadriaticum and the coccolithophore E. huxleyi. Thus, at least these three species need concerted future efforts.
The combination of results presented herein together with previously published protocols from the EMS initiative50 significantly expands the segment of extant eukaryotic diversity amenable to reverse genetics approaches. Out of the 39 microbial eukaryotes selected for the initiative, exogenous DNA was delivered and expressed in more than 50% of them. The transformation systems enable us to shed light on the function of species-specific genes, which likely reflect key adaptations to specific niches in dynamic ocean habitats.
Methods
Studied species and used transformation methods
For the full list of vector sequences and maps see Supplementary Notes 1 and for detailed description of Figs. 3 and 4 see Supplementary Note 2. Antibiotic concentrations effective for selection of transformants can be found in Supplementary Table 3, the details of the transformation methods applied to this study in Supplementary Table 4 and contact details for individual laboratories in Supplementary Table 6. Full list of protists (including details of culture collection) and links to the complete step-by-step transformation protocols and published vector sequences are listed in Supplementary Table 5. The protocols.io links listed in Table 1 and Supplementary Table 5 are summarized in Supplementary Tables 7 and 8.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The data that support the findings of this study are available from the corresponding authors as well as the other authors upon request (for the contacts see Supplementary Table 6). Source data for Figs. 3 and 4 and Supplementary Figs. 9b,c, 11a and 12b,c are available online.
Change history
15 April 2020
A Correction to this paper has been published: https://doi.org/10.1038/s41592-020-0828-6
References
- Worden, A. Z. et al. Rethinking the marine carbon cycle: factoring in the multifarious lifestyles of microbes. Science 347, 1257594 (2015).
PubMed Google Scholar - de Vargas, C. et al. Eukaryotic plankton diversity in the sunlit global ocean. Science 348, 1261605 (2015).
PubMed Google Scholar - Collier, J. L. & Rest, J. S. Swimming, gliding, and rolling toward the mainstream: cell biology of marine protists. Mol. Biol. Cell. 30, 1245–1248 (2019).
CAS PubMed PubMed Central Google Scholar - Curtis, B. A. et al. Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature 492, 59–65 (2012).
CAS PubMed Google Scholar - Armbrust, E. V. et al. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79–86 (2004).
CAS PubMed Google Scholar - Read, B. A. et al. Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature 499, 209–213 (2013).
CAS PubMed Google Scholar - Keeling, P. J. et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 12, e1001889 (2014).
PubMed PubMed Central Google Scholar - Nymark, M., Sharma, A. K., Sparstad, T., Bones, A. M. & Winge, P. A. CRISPR/Cas9 system adapted for gene editing in marine algae. Sci. Rep. 6, 24951 (2016).
CAS PubMed PubMed Central Google Scholar - Hopes, A., Nekrasov, V., Kamoun, S. & Mock, T. Editing of the urease gene by CRISPR-Cas in the diatom Thalassiosira pseudonana. Plant Meth. 12, 49 (2016).
Google Scholar - Carradec, Q. et al. A global atlas of eukaryotic genes. Nat. Commun. 9, 373 (2018).
PubMed PubMed Central Google Scholar - Apt, K. E., Kroth-Pancic, P. G. & Grossman, A. R. Stable nuclear transformation of the diatom Phaeodactylum tricornutum. Mol. Gen. Genet. 252, 572–579 (1996).
CAS PubMed Google Scholar - Paschke, P. et al. Genetic engineering of Dictyostelium discoideum cells based on selection and growth on bacteria. J. Vis. Exp. 143, e58981 (2019).
Google Scholar - Karas, B. J. et al. Designer diatom episomes delivered by bacterial conjugation. Nat. Commun. 6, 6925 (2015).
CAS PubMed Google Scholar - Hirakawa, Y., Kofuji, R. & Ishida, K. Transient transformation of a chlorarachniophyte alga, Lotharella amoebiformis (chlorarachniophyceae), with uidA and egfp reporter genes. J. Phycol. 44, 814–820 (2008).
CAS PubMed Google Scholar - Nimmo, I. C. et al. Genetic transformation of the dinoflagellate chloroplast. eLife 8, e45292 (2019).
PubMed PubMed Central Google Scholar - Fernández Robledo, J. A., Lin, Z. & Vasta, G. R. Transfection of the protozoan parasite Perkinsus marinus. Mol. Biochem. Parasitol. 157, 44–53 (2008).
PubMed Google Scholar - Sakamoto, H. et al. Puromycin selection for stable transfectants of the oyster-infecting parasite Perkinsus marinus. Parasitol. Int. 69, 13–16 (2018).
PubMed Google Scholar - Kaur, B. et al. Transformation of Diplonema papillatum, the type species of the highly diverse and abundant marine micro-eukaryotes Diplonemida (Euglenozoa). Env. Microbiol. 20, 1030–1040 (2018).
CAS Google Scholar - Diao, J., Song, X., Zhang, X., Chen, L. & Zhang, W. Genetic engineering of Crypthecodinium cohnii to increase growth and lipid accumulation. Front. Microbiol. 9, 492 (2018).
PubMed PubMed Central Google Scholar - Sakaguchi, K. et al. Versatile transformation system that is applicable to both multiple transgene expression and gene targeting for thraustochytrids. Appl. Environ. Microbiol. 78, 3193–3202 (2012).
CAS PubMed PubMed Central Google Scholar - Booth, D., Middleton, H. & King, N. Choanoflagellate transfection illuminates their cell biology and the ancestry of animal septins. Mol. Biol. Cell. 29, 3026–3038 (2018).
CAS PubMed PubMed Central Google Scholar - Wetzel, L. A. et al. Predicted glycosyltransferases promote development and prevent spurious cell clumping in the choanoflagellate S. rosetta. eLife 7, e41482 (2018).
PubMed PubMed Central Google Scholar - van Baren, M. J. et al. Evidence-based green algal genomics reveals marine diversity and ancestral characteristics of land plants. BMC Genomics 17, 1–22 (2016).
Google Scholar - Lozano, J. C. et al. Efficient gene targeting and removal of foreign DNA by homologous recombination in the picoeukaryote Ostreococcus. Plant J. 78, 1073–1083 (2014).
CAS PubMed Google Scholar - Van Ooijen, G., Knox, K., Kis, K., Bouget, F. Y. & Millar, A. J. Genomic transformation of the picoeukaryote Ostreococcus tauri. J. Vis. Exp. 65, e4074 (2012).
Google Scholar - Hovde, B. T. et al. Genome sequence and transcriptome analyses of Chrysochromulina tobin: metabolic tools for enhanced algal fitness in the prominent order Prymnesiales (Haptophyceae). PLoS Genet. 11, e1005469 (2015).
PubMed PubMed Central Google Scholar - Endo, H. et al. Stable nuclear transformation system for the coccolithophorid alga Pleurochrysis carterae. Sci. Rep. 6, 22252 (2016).
CAS PubMed PubMed Central Google Scholar - Dörner, J., Carbonell, P., Pino, S. & Farias, A. Variation of fatty acids in Isochrysis galbana (T-Iso) and Tetraselmis suecica, cultured under different nitrate availabilities. J. Fish. Aquacult. 5, 1–3 (2014).
Google Scholar - Dunahay, T. G., Jarvis, E. E. & Roessler, P. G. Genetic transformation of the diatoms Cyclotella cryptica and Navicula saprophila. J. Phycol. 31, 1004–1012 (1995).
CAS Google Scholar - Field, C. B., Behrenfeld, M. J., Randerson, J. T. & Falkowski, P. Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281, 237–240 (1998).
CAS PubMed Google Scholar - Mishra, M., Arukha, A. P., Bashir, T., Yadav, D. & Prasad, G. B. K. S. All new faces of diatoms: potential source of nanomaterials and beyond. Front. Microbiol. 8, 1239 (2017).
PubMed PubMed Central Google Scholar - Mock, T. et al. Evolutionary genomics of the cold-adapted diatom Fragilariopsis cylindrus. Nature 541, 536–540 (2017).
CAS PubMed Google Scholar - Brunson, J. K. et al. Biosynthesis of the neurotoxin domoic acid in a bloom-forming diatom. Science 361, 1356–1358 (2018).
CAS PubMed PubMed Central Google Scholar - Kroth, P. G. Genetic transformation: a tool to study protein targeting in diatoms. Methods Mol. Biol. 390, 257–267 (2007).
CAS PubMed Google Scholar - Sabatino, V. et al. Establishment of genetic transformation in the sexually reproducing diatoms _Pseudo_-nitzschia multistriata and Pseudo-nitzschia arenysensis and inheritance of the transgene. Mar. Biotech. 17, 452–462 (2015).
CAS Google Scholar - Ono, K., Aki, T. & Kawamoto, S. Method for introducing a gene into labyrinthulomycota. US patent 7,888,123 (2011).
- Kilian, O., Benemann, C. S., Niyogi, K. K. & Vick, B. High-efficiency homologous recombination in the oil-producing alga Nannochloropsis sp. Proc. Natl Acad. Sci. USA 108, 21265–21269 (2011).
CAS PubMed PubMed Central Google Scholar - Duda, K. et al. High-efficiency genome editing via 2A-coupled co-expression of fluorescent proteins and zinc finger nucleases of CRISPR/Cas9 nicase pairs. Nucl. Acids Res. 42, e84 (2014).
CAS PubMed PubMed Central Google Scholar - Donald, R. G. & Roos, D. S. Stable molecular transformation of Toxoplasma gondii: a selectable dihydrofolate reductase-thymidylate synthase marker based on drug-resistance mutations in malaria. Proc. Natl Acad. Sci. USA 90, 11703–11707 (1993).
CAS PubMed PubMed Central Google Scholar - Barbrook, A. C., Howe, C. J. & Nisbet, R. E. R. Breaking up is hard to do: the complexity of the dinoflagellate chloroplast genome. Perspect. Phycol. 6, 31–37 (2019).
Google Scholar - Sprecher, B. N., Zhang, H. & Lin, S. Nuclear gene transformation in the dinoflagellate Oxyrrhis marina. Microorganisms 8, 126 (2020).
CAS PubMed Central Google Scholar - Zhang, H. et al. Signal recognition particle RNA in dinoflagellates and the perkinsid Perkinsus marinus. Protist 164, 748–761 (2013).
CAS PubMed Google Scholar - Chambouvet, A. et al. Cryptic infection of a broad taxonomic and geographic diversity of tadpoles by Perkinsea protists. Proc. Natl Acad. Sci. USA 112, E4743–E4751 (2015).
CAS PubMed PubMed Central Google Scholar - Adl, S. M. et al. Revision to the classification, nomenclature and diversity of eukaryotes. J. Euk. Microbiol. 66, 4–119 (2019).
PubMed Google Scholar - Matthews, K. R. 25 years of African trypanosome research: from description to molecular dissection and new drug discovery. Mol. Biochem. Parasitol. 200, 30–40 (2015).
CAS PubMed PubMed Central Google Scholar - Opperdoes, F. R., Butenko, A., Flegontov, P., Yurchenko, V. & Lukeš, J. Comparative metabolism of free-living Bodo saltans and parasitic trypanosomatids. J. Eukaryot. Microbiol. 63, 657–678 (2016).
CAS PubMed Google Scholar - Richter, D. J., Fozouni, P., Eisen, M. & King, N. Gene family innovation, conservation and loss on the animal stem lineage. eLife 7, 1–43 (2018).
Google Scholar - Parra-Acero, H. et al. Transfection of Capsaspora owczarzaki, a close unicellular relative of animals. Development 145, 162107 (2018).
Google Scholar - Suga, H. & Ruiz-Trillo, I. Development of ichthyosporean sheds light on the origin of metazoan multicellularity. Dev. Biol. 377, 284–292 (2013).
CAS PubMed PubMed Central Google Scholar - Waller, R. F. et al. Strength in numbers: collaborative science for new experimental model systems. PLoS Biol. 16, e2006333 (2018).
PubMed PubMed Central Google Scholar
Acknowledgements
We thank M. Salisbury and D. Lacono, C. Poirier, M. Hamilton, C. Eckmann, H. Igel, C. Yung and K. Hoadley for assistance; V.K. Nagarajan, M. Accerbi and P.J. Green who carried out Agrobacterium studies in Heterosigma akashiwo, and N. Kraeva, C. Bianchi and V. Yurchenko for the help with designing the p57-V5+NeoR construct. We are also grateful to the protocols.io team (L. Teytelman and A. Broellochs) for their support. This collaborative effort was supported by the Gordon and Betty Moore Foundation EMS Program of the Marine Microbiology Initiative (grant nos. GBMF4972 and 4972.01 to F.-Y.B.; GBMF4970 and 4970.01 to M.A. and A.Z.W.; GBMF3788 to A.Z.W.; GBMF 4968 and 4968.01 to H.C.; GBMF4984 to V.H.; GBMF4974 and 4974.01 to C. Brownlee; GBMF4964 to Y. Hirakawa; GBMF4961 to T. Mock; GBMF4958 to P.S.; GBMF4957 to A.T.; GBMF4960 to G.J.S.; GBMF4979 to K.C.; GBMF4982 and 4982.01 to J.L.C.; GBMF4964 to P.J.K.; GBMF4981 to P.v.D.; GBMF5006 to A.E.A.; GBMF4986 to C.M.; GBMF4962 to J.A.F.R.; GBMF4980 and 4980.01 to S.L.; GBMF 4977 and 4977.01 to R.F.W.; GBMF4962.01 to C.H.S.; GBMF4985 to J.M.; GBMF4976 and 4976.01 to C.H.; GBMF4963 and 4963.01 to V.E.; GBMF5007 to C.L.D.; GBMF4983 and 4983.01 to J.L.; GBMF4975 and 4975.01 to A.D.T.; GBMF4973 and 4973.01 to I.R.-T. and GBMF4965 to N.K.), by The Leverhulme Trust (RPG-2017-364) to T. Mock and A. Hopes, and by ERD funds (16_019/0000759) from the Czech Ministry of Education to J.L.
Author information
Author notes
- R. Ellen R. Nisbet
Present address: School of Biosciences, University of Nottingham, Sutton Bonington, UK - Pía A. Elustondo
Present address: AGADA Biosciences Inc., Halifax, Nova Scotia, Canada - Luke Noble
Present address: Institute de Biologie de l’ENS, Département de biologie, École Normale Supérieure, CNRS, INSERM, Paris, France - Albane Ruaud & Estienne C. Swart
Present address: Max Planck Institute for Developmental Biology, Tübingen, Germany - These authors contributed equally: Drahomíra Faktorová, R. Ellen R. Nisbet, José A. Fernández Robledo, Elena Casacuberta, Lisa Sudek.
Authors and Affiliations
- Institute of Parasitology, Biology Centre, Czech Academy of Sciences and Faculty of Sciences, University of South Bohemia, České Budějovice, Czech Republic
Drahomíra Faktorová, Ambar Kachale, Binnypreet Kaur & Julius Lukeš - Department of Biochemistry, University of Cambridge, Cambridge, UK
R. Ellen R. Nisbet, Adrian C. Barbrook, Elin Einarsson, Christopher J. Howe, Ian Hu, Imen Lassadi, Isabel C. Nimmo & Ross F. Waller - Bigelow Laboratory for Ocean Sciences, East Boothbay, ME, USA
José A. Fernández Robledo, Duncan B. Coles & Nastasia J. Freyria - Institut de Biologia Evolutiva, CSIC-Universitat Pompeu Fabra, Barcelona, Spain
Elena Casacuberta, Cristina Aresté, Sebastián R. Najle & Iñaki Ruiz-Trillo - Monterey Bay Aquarium Research Institute, Moss Landing, CA, USA
Lisa Sudek, Jian Guo & Alexandra Z. Worden - Integrative Oceanography Division, Scripps Institution of Oceanography, University of California, San Diego, CA, USA
Andrew E. Allen, Mark Moosburner & Jernej Turnšek - Microbial and Environmental Genomics, J. Craig Venter Institute, La Jolla, CA, USA
Andrew E. Allen, Christopher L. Dupont, Mark Moosburner & Jernej Turnšek - Molecular, Cell and Developmental Biology, University of California, Santa Cruz, CA, USA
Manuel Ares Jr & Jian Guo - The Marine Biological Association, Plymouth and School of Ocean and Earth Sciences, University of Southampton, Southampton, UK
Cecilia Balestreri, Colin Brownlee, Andrea Highfield, Rowena Stern & Glen Wheeler - School of Biological Sciences, University of Nebraska, Lincoln, NE, USA
Patrick Beardslee, Heriberto Cerutti, Thomas Clemente, Fulei Luan & Xiaoxue Wen - Gordon and Betty Moore Foundation, Palo Alto, CA, USA
Sara Bender, Adam C. Jones & Jonathan Z. Kaye - Department of Molecular and Cell Biology, University of California, Berkeley, CA, USA
David S. Booth, Nicole King & Monika A. Sigg - Sorbonne Université, CNRS UMR7621, Observatoire Océanologique, Banyuls sur Mer, France
François-Yves Bouget, Jean-Claude Lozano & Valérie Vergé - Institut de Biologie de l’Ecole Normale Supérieure (IBENS), Ecole Normale Supérieure, CNRS, INSERM, Université PSL, Paris, France
Chris Bowler - Centre for Comparative Genomics and Evolutionary Bioinformatics, Dalhousie University, Halifax, Nova Scotia, Canada
Susana A. Breglia, Pía A. Elustondo, Esteban R. Haro-Contreras & Claudio H. Slamovits - Department of Biochemistry and Robert-Cedergren Centre for Bioinformatics and Genomics, Université de Montréal, Montreal, Quebec, Canada
Gertraud Burger & Matus Valach - Institute of Cell Biology, University of Bern, Bern, Switzerland
Rachele Cesaroni, Mariusz Nowacki & Estienne C. Swart - Center for Tropical and Emerging Global Diseases, University of Georgia, Athens, GA, USA
Miguel A. Chiurillo, Roberto Docampo, Noelia Lander & Zhuhong Li - School of Marine and Atmospheric Sciences, Stony Brook University, Stony Brook, NY, USA
Jackie L. Collier & Mariana Rius - Department of Botany, University of British Columbia, Vancouver, British Columbia, Canada
Elizabeth C. Cooney, Elisabeth Hehenberger, Nicholas A. T. Irwin & Patrick J. Keeling - University of Delaware College of Earth, Ocean and Environment, Lewes, DE, USA
Kathryn Coyne & Deepak Nanjappa - Woods Hole Oceanographic Institution, Woods Hole, MA, USA
Virginia Edgcomb - Facultad Ciencias Biológicas, Pontificia Universidad Católica de Chile, Fondo de Desarrollo de Areas Prioritarias, Center for Genome Regulation and Millennium Institute for Integrative Biology (iBio), Santiago de Chile, Chile
Fernan Federici, Jorge Ibañez, Tamara Matute, Isaac Nuñez, Albane Ruaud & Peter von Dassow - School of Biosciences, University of Kent, Canterbury, Kent, UK
Veronica Freire-Beneitez, Eleanna Kazana, Jan Pyrih, Anastasios D. Tsaousis & Tobias von der Haar - Laboratory of Molecular and Evolutionary Parasitology, University of Kent, Kent, UK
Veronica Freire-Beneitez, Eleanna Kazana & Anastasios D. Tsaousis - Graduate School of Life and Environmental Sciences, University of Tsukuba, Ibaraki, Japan
Kodai Fukuda - Department of Mechanical Engineering, Massachusetts Institute of Technology, Boston, MA, USA
Paulo A. García - Department of Organismic and Evolutionary Biology, Harvard University, Cambridge, MA, USA
Peter R. Girguis & Fatma Gomaa - Centre for Organismal Studies, University of Heidelberg, Heidelberg, Germany
Sebastian G. Gornik - Department of Parasitology, Faculty of Science, Charles University, BIOCEV, Vestec, Czech Republic
Vladimír Hampl, Natalia Ewa Janowicz, Anna M. G. Novák Vanclová & Jan Pyrih - Faculty of Life and Environmental Sciences, University of Tsukuba, Ibaraki, Japan
Yutaka Hanawa & Yoshihisa Hirakawa - School of Environmental Sciences, University of East Anglia, Norwich, UK
Amanda Hopes & Thomas Mock - Graduate School of Life Sciences, Tohoku University, Sendai, Miyagi, Japan
Yuu Ishii & Shinichiro Maruyama - Division of Environmental Photobiology, National Institute for Basic Biology, Okazaki, Aichi, Japan
Konomi Fujimura-Kamada & Jun Minagawa - Department of Marine Sciences, University of Connecticut , Groton, CT, USA
Lawrence A. Klobutcher, Senjie Lin, Brittany N. Sprecher & Huan Zhang - School of Biosciences and Veterinary Medicine, University of Camerino, Camerino, Italy
Cristina Miceli, Angela Piersanti & Sandra Pucciarelli - Department of Basic Biology, School of Life Science, Graduate University for Advanced Studies, Okazaki, Aichi, Japan
Jun Minagawa - Instituto de Biología Molecular y Celular, CONICET, and Facultad de Ciencias Bioquímicas y Farmacéuticas, Universidad Nacional de Rosario, Rosario, Argentina
Sebastián R. Najle - Center for Genomics and Systems Biology, New York University, New York, NY, USA
Luke Noble - Biological and Environmental Science and Engineering Division, King Abdullah University of Science and Technology, Thuwal, Saudi Arabia
Arnab Pain - Center for Zoonosis Control, Global Institution for Collaborative Research and Education, Hokkaido University, Sapporo, Japan
Arnab Pain - Department of Ecology and Evolution, Stony Brook University, Stony Brook, NY, USA
Joshua S. Rest - Lasry Center for Biosciences, Clark University, Worcester, MA, USA
Deborah Robertson - Departament de Genètica Microbiologia i Estadıśtica, Universitat de Barcelona, Barcelona, Spain
Iñaki Ruiz-Trillo - Catalan Institution for Research and Advanced Studies, Barcelona, Spain
Iñaki Ruiz-Trillo - Department of Systems Biology, Harvard Medical School, Boston, MA, USA
Pamela A. Silver & Jernej Turnšek - Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA, USA
Pamela A. Silver & Jernej Turnšek - Department of Environmental Biotechnology, Moss Landing Marine Laboratories, Moss Landing, CA, USA
G. Jason Smith & April Woods - Department of Molecular Genetics and Cell Biology, University of Chicago, Chicago, IL, USA
Lev Tsypin & Aaron Turkewitz - Department of Biology, California Institute of Technology, Pasadena, CA, USA
Lev Tsypin - Instituto Milenio de Oceanografia de Chile, Concepción, Chile
Peter von Dassow - Institute of Oceanography, Minjiang University, Fuzhou, China
Lu Wang - Ocean EcoSystems Biology Unit, Marine Ecology Division, Helmholtz Centre for Ocean Research, Kiel, Germany
Alexandra Z. Worden
Authors
- Drahomíra Faktorová
You can also search for this author inPubMed Google Scholar - R. Ellen R. Nisbet
You can also search for this author inPubMed Google Scholar - José A. Fernández Robledo
You can also search for this author inPubMed Google Scholar - Elena Casacuberta
You can also search for this author inPubMed Google Scholar - Lisa Sudek
You can also search for this author inPubMed Google Scholar - Andrew E. Allen
You can also search for this author inPubMed Google Scholar - Manuel Ares Jr
You can also search for this author inPubMed Google Scholar - Cristina Aresté
You can also search for this author inPubMed Google Scholar - Cecilia Balestreri
You can also search for this author inPubMed Google Scholar - Adrian C. Barbrook
You can also search for this author inPubMed Google Scholar - Patrick Beardslee
You can also search for this author inPubMed Google Scholar - Sara Bender
You can also search for this author inPubMed Google Scholar - David S. Booth
You can also search for this author inPubMed Google Scholar - François-Yves Bouget
You can also search for this author inPubMed Google Scholar - Chris Bowler
You can also search for this author inPubMed Google Scholar - Susana A. Breglia
You can also search for this author inPubMed Google Scholar - Colin Brownlee
You can also search for this author inPubMed Google Scholar - Gertraud Burger
You can also search for this author inPubMed Google Scholar - Heriberto Cerutti
You can also search for this author inPubMed Google Scholar - Rachele Cesaroni
You can also search for this author inPubMed Google Scholar - Miguel A. Chiurillo
You can also search for this author inPubMed Google Scholar - Thomas Clemente
You can also search for this author inPubMed Google Scholar - Duncan B. Coles
You can also search for this author inPubMed Google Scholar - Jackie L. Collier
You can also search for this author inPubMed Google Scholar - Elizabeth C. Cooney
You can also search for this author inPubMed Google Scholar - Kathryn Coyne
You can also search for this author inPubMed Google Scholar - Roberto Docampo
You can also search for this author inPubMed Google Scholar - Christopher L. Dupont
You can also search for this author inPubMed Google Scholar - Virginia Edgcomb
You can also search for this author inPubMed Google Scholar - Pía A. Elustondo
You can also search for this author inPubMed Google Scholar - Fernan Federici
You can also search for this author inPubMed Google Scholar - Veronica Freire-Beneitez
You can also search for this author inPubMed Google Scholar - Nastasia J. Freyria
You can also search for this author inPubMed Google Scholar - Kodai Fukuda
You can also search for this author inPubMed Google Scholar - Paulo A. García
You can also search for this author inPubMed Google Scholar - Peter R. Girguis
You can also search for this author inPubMed Google Scholar - Fatma Gomaa
You can also search for this author inPubMed Google Scholar - Sebastian G. Gornik
You can also search for this author inPubMed Google Scholar - Jian Guo
You can also search for this author inPubMed Google Scholar - Vladimír Hampl
You can also search for this author inPubMed Google Scholar - Yutaka Hanawa
You can also search for this author inPubMed Google Scholar - Esteban R. Haro-Contreras
You can also search for this author inPubMed Google Scholar - Elisabeth Hehenberger
You can also search for this author inPubMed Google Scholar - Andrea Highfield
You can also search for this author inPubMed Google Scholar - Yoshihisa Hirakawa
You can also search for this author inPubMed Google Scholar - Amanda Hopes
You can also search for this author inPubMed Google Scholar - Christopher J. Howe
You can also search for this author inPubMed Google Scholar - Ian Hu
You can also search for this author inPubMed Google Scholar - Jorge Ibañez
You can also search for this author inPubMed Google Scholar - Nicholas A. T. Irwin
You can also search for this author inPubMed Google Scholar - Yuu Ishii
You can also search for this author inPubMed Google Scholar - Natalia Ewa Janowicz
You can also search for this author inPubMed Google Scholar - Adam C. Jones
You can also search for this author inPubMed Google Scholar - Ambar Kachale
You can also search for this author inPubMed Google Scholar - Konomi Fujimura-Kamada
You can also search for this author inPubMed Google Scholar - Binnypreet Kaur
You can also search for this author inPubMed Google Scholar - Jonathan Z. Kaye
You can also search for this author inPubMed Google Scholar - Eleanna Kazana
You can also search for this author inPubMed Google Scholar - Patrick J. Keeling
You can also search for this author inPubMed Google Scholar - Nicole King
You can also search for this author inPubMed Google Scholar - Lawrence A. Klobutcher
You can also search for this author inPubMed Google Scholar - Noelia Lander
You can also search for this author inPubMed Google Scholar - Imen Lassadi
You can also search for this author inPubMed Google Scholar - Zhuhong Li
You can also search for this author inPubMed Google Scholar - Senjie Lin
You can also search for this author inPubMed Google Scholar - Jean-Claude Lozano
You can also search for this author inPubMed Google Scholar - Fulei Luan
You can also search for this author inPubMed Google Scholar - Shinichiro Maruyama
You can also search for this author inPubMed Google Scholar - Tamara Matute
You can also search for this author inPubMed Google Scholar - Cristina Miceli
You can also search for this author inPubMed Google Scholar - Jun Minagawa
You can also search for this author inPubMed Google Scholar - Mark Moosburner
You can also search for this author inPubMed Google Scholar - Sebastián R. Najle
You can also search for this author inPubMed Google Scholar - Deepak Nanjappa
You can also search for this author inPubMed Google Scholar - Isabel C. Nimmo
You can also search for this author inPubMed Google Scholar - Luke Noble
You can also search for this author inPubMed Google Scholar - Anna M. G. Novák Vanclová
You can also search for this author inPubMed Google Scholar - Mariusz Nowacki
You can also search for this author inPubMed Google Scholar - Isaac Nuñez
You can also search for this author inPubMed Google Scholar - Arnab Pain
You can also search for this author inPubMed Google Scholar - Angela Piersanti
You can also search for this author inPubMed Google Scholar - Sandra Pucciarelli
You can also search for this author inPubMed Google Scholar - Jan Pyrih
You can also search for this author inPubMed Google Scholar - Joshua S. Rest
You can also search for this author inPubMed Google Scholar - Mariana Rius
You can also search for this author inPubMed Google Scholar - Deborah Robertson
You can also search for this author inPubMed Google Scholar - Albane Ruaud
You can also search for this author inPubMed Google Scholar - Iñaki Ruiz-Trillo
You can also search for this author inPubMed Google Scholar - Monika A. Sigg
You can also search for this author inPubMed Google Scholar - Pamela A. Silver
You can also search for this author inPubMed Google Scholar - Claudio H. Slamovits
You can also search for this author inPubMed Google Scholar - G. Jason Smith
You can also search for this author inPubMed Google Scholar - Brittany N. Sprecher
You can also search for this author inPubMed Google Scholar - Rowena Stern
You can also search for this author inPubMed Google Scholar - Estienne C. Swart
You can also search for this author inPubMed Google Scholar - Anastasios D. Tsaousis
You can also search for this author inPubMed Google Scholar - Lev Tsypin
You can also search for this author inPubMed Google Scholar - Aaron Turkewitz
You can also search for this author inPubMed Google Scholar - Jernej Turnšek
You can also search for this author inPubMed Google Scholar - Matus Valach
You can also search for this author inPubMed Google Scholar - Valérie Vergé
You can also search for this author inPubMed Google Scholar - Peter von Dassow
You can also search for this author inPubMed Google Scholar - Tobias von der Haar
You can also search for this author inPubMed Google Scholar - Ross F. Waller
You can also search for this author inPubMed Google Scholar - Lu Wang
You can also search for this author inPubMed Google Scholar - Xiaoxue Wen
You can also search for this author inPubMed Google Scholar - Glen Wheeler
You can also search for this author inPubMed Google Scholar - April Woods
You can also search for this author inPubMed Google Scholar - Huan Zhang
You can also search for this author inPubMed Google Scholar - Thomas Mock
You can also search for this author inPubMed Google Scholar - Alexandra Z. Worden
You can also search for this author inPubMed Google Scholar - Julius Lukeš
You can also search for this author inPubMed Google Scholar
Contributions
The project was conceived and designed by A.C.J., J.Z.K., S.B., D.F., J.L., R.E.R.N., J.A.F.R., E.C., L.S., A.Z.W., T. Mock, A.E.A., F.-Y.B, C. Brownlee, C. Bowler, H.C., T.C., J.L.C., K.C., C.L.D., V.E., V.H., Y. Hirakawa, C.J.H., P.J.K., N.K., S.L., C.M., J.M., I.R.-T., P.A.S., C.H.S., G.J.S., A.D.T., P.v.D., A.T. and R.F.W. Data analysis was carried out by M.A.Jr, C.A., C. Balestreri, A.C.B., P.B., D.S.B., S.A.B., G.B., R.C., M.A.C., D.B.C., E.C.C., R.D., E.E., P.A.E., F.F., V.F.-B., N.J.F., K.F., P.A.G., P.R.G., F.G., S.G.G., J.G., Y. Hanawa, E.R.H.-C., E.H., A. Highfield, A. Hopes, I.H., J.I., N.A.T.I., Y.I., N.E.J., A.K., K.F.-K., B.K., E.K., L.A.K., N.L., I.L., Z.L., J.-C.L., F.L., S.M., T. Matute, M.M., S.R.N., D.N., I.C.N., L.N., A.M.G.N.V., M.N., I.N., A. Pain, A. Piersanti, S.P., J.P., J.S.R., M.R., D.R., A.R., M.A.S., E.C.S., B.N.S., R.S., T.v.d.H., L.T., J.T., M.V., V.V., L.W., X.W., G.W., A.W. and H.Z. The manuscript was written by D.F., R.E.R.N., J.A.F.R., E.C., L.S., T. Mock, A.Z.W. and J.L. with input from all authors.
Corresponding authors
Correspondence toDrahomíra Faktorová, Thomas Mock, Alexandra Z. Worden or Julius Lukeš.
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Lei Tang was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Faktorová, D., Nisbet, R.E.R., Fernández Robledo, J.A. et al. Genetic tool development in marine protists: emerging model organisms for experimental cell biology.Nat Methods 17, 481–494 (2020). https://doi.org/10.1038/s41592-020-0796-x
- Received: 26 June 2019
- Accepted: 02 March 2020
- Published: 06 April 2020
- Issue Date: 02 May 2020
- DOI: https://doi.org/10.1038/s41592-020-0796-x