Death and more: DNA damage response pathways in the nematode C. elegans (original) (raw)
Introduction
Living organisms expend considerable energy to preserve integrity of their genomes. Multiple mechanisms have evolved to ensure the fidelity of genome duplication and to guarantee faithful partitioning of chromosomes at each cell division. Furthermore, cells are under the constant threat of DNA damaging agents, such as ionizing radiation, oxygen radicals and monofunctional alkylating agents. Maintenance of genome stability thus also depends on an appropriate response when DNA damage is inflicted by these agents. Indeed, eukaryotes have developed complex biochemical signaling networks that activate numerous processes following DNA damage: activation of the repair machinery, transient cell cycle arrest, transcriptional upregulation of a number of other response proteins and in metazoans, induction of apoptosis. Together, these response pathways insure efficient repair of the lesion or, if necessary, elimination of the damaged cell. Much of our understanding of DNA damage response pathways originate from elegant genetic studies in the yeasts Saccharomyces cerevisiae and S. pombe. These studies, together with work performed in Aspergillus, Caenorhabditis elegans, Drosophila, various mammalian species including humans, have revealed much conservation, but also some surprising changes in DNA damage response pathways through evolution.
In this review, we very briefly discuss the general features of DNA damage response pathways and their main players, and then concentrate on what has been learned about DNA damage response from studies in the nematode C. elegans.
DNA Damage Response Proteins: Sensors, Transducers and Effectors
Depending on their distinct positions and functions within the signaling cascades, proteins involved in DNA damage response have been classified as sensors that detect the damage, transducers that transmit the signal that damage is present, or effectors that elicit the various specific biological responses (Table 1).
Table 1 Orthologous checkpoint proteins in C. elegans, yeasts and mammalian cells
Sensor proteins are thought to associate with damaged DNA directly or indirectly and they serve as recognition complexes to recruit and modulate the function of specific transducer proteins. Different types of DNA damage can activate different molecular pathways, suggesting that the nature of the DNA structure that is recognized by the sensors defines each time the steps to follow. However, despite the large number of possible DNA lesion types, only a limited number of response pathways have been identified. It is likely that the DNA damage response pathways sense common intermediates, rather than the original damage. Indeed, the current view for eukaryotic cells is that all types of DNA damage are eventually converted to either single-strand (ssDNA) and double-strand DNA breaks (DSBs).
Two complexes – the Rad17-RFC and the 9-1-1 complexes – cooperate to detect DSBs. Rad17 interacts with four replication factor C subunits (Rfc2, Rfc3, Rfc4, Rfc5) to form a pentameric structure,1,2 whereas the proliferating cell nuclear antigen (PCNA) homologs Rad9, hydroxyurea-sensitive 1 (Hus1) and Rad1 form a heterotrimeric ring around DNA. These two complexes are thus reminiscent of the ‘clamp loader’ and the ‘sliding clamp’ involved in DNA replication,3 and recent evidence suggests that a similar mechanism might be used to load the 9-1-1 complex onto sites of DNA damage.4 A third protein complex, known as the Mre11–Rad50–Nbs1 complex (MRN), Mre11=meiotic recombination 11; Nbs1 nibrin (mutated in Nijmegen breakage syndrome), also localizes to sites of double-strand breaks; this complex furthermore has a role in repair, meiotic recombination and telomere maintenance. Mutants for the Mre11 or Nbs1 proteins exhibit a radioresistant DNA synthesis (RDS) phenotype, indicating that these proteins are also involved in checkpoints during S-phase of the cell cycle.5,6,7 Finally, breast cancer 1 (BRCA1), which likely acts as an adaptor molecule and colocalizes with proteins such as Rad51, the PCNA and MRN complexes, plays an important role in several distinct DNA damage response pathways.8,9,10 Mutations in the BRCA1 gene are associated with more than half of all cases of familial breast cancer, underscoring the importance of genome integrity in protecting against cancer development.
Downstream of the sensor molecules, transducers of the initial signal initiate phosphorylation cascades that amplify and diversify the signal by targeting multiple downstream effectors. This class of proteins includes two prominent members of the PI3 K superfamily (phosphatidyl-inositol-3-kinase), ATM and ATR (the homologs of S. pombe Tel1 and Rad3, respectively) (ATM=active telangiectasia mutated kinase; ATR=ataxia telangiectasia-related kinase; Tel1=telomere length regulation 1). Upon their activation by DNA damage, ATM and/or ATR phosphorylate a number of substrates, whose identity can vary according to the nature of the lesion. DNA damage responses are highly abnormal in cells lacking ATM or ATR, leading to accumulation of mutations and chromosomal aberrations, which increase the probability of developmental abnormalities and genetic diseases.11,12,13,14 The serine–threonine kinases checkpoint kinase 1 (Chk1) and Chk2 are among the phosphorylated substrates and the molecules that will carry on the signal, and are required for cell cycle arrest following DNA damage.12,14,15
Cell Cycle Arrest
DNA damage temporarily arrests cell cycle progression, in order to permit repair prior to DNA replication or cell division. The presence of eukaryotic cell cycle checkpoints that respond to DNA damage were first inferred from the identification of radiation-sensitive (rad) yeast mutants that fail to delay entry into mitosis after DNA damage. Subsequently, checkpoint pathways have also been identified that control entry into or progression through S phase.
The G1/S checkpoint
When damage occurs in the G1 phase, most eukaryotic cells exhibit a delay prior to S-phase onset. This prevents replication of a damaged template that might result in the fixation of mutations or in a chromosome bearing a DSB. Whereas this checkpoint is weak and most of the damage remains unrepaired in budding yeast,16 the G1/S checkpoint is prominent in mammals, where it acts by preventing activation of Cdk2-cyclin E (Cdk=cyclin-dependent kinase). This is accomplished by stabilizing p53 through phosphorylation to cause transcriptional activation of p21 and by degrading Cdc25A (Cds=cell division control) to maintain the inhibitory phosphorylation on Cdk2.17,18
The intra-S-phase checkpoint
Most of what is known about this checkpoint control comes from studies in budding and fission yeasts, with mammalian cells showing features similar to the latter. Upon encountering a DNA lesion, the replication forks stall transiently to block early- and late-firing onsets. Forks are then converted to structures that are prevented from undergoing nuclease attack and collapse. Those that are processed by nucleases activate the ATR homolog, Rad3, and Cds1/Chk2 activity. Inhibition of CDK2 activity through Cdc25A degradation in mammalian cells leads to a several-hour delay in S-phase progression, whereas prolonged blockade in yeast may cause regaining of replication.19
The G2/M checkpoint
In both yeast and higher eukaryotes, the G2/M transition is blocked by the maintenance of the inhibitory phosphorylation on Cdc2, and thus blockade of Cdc2-cyclin B activity. Activation of ATR or ATM by DNA damage leads to Chk1 or Chk2 activation, respectively which, in turn, act on Cdc25C phosphatase to promote its association with 14-3-3 proteins.20,21,22 Additionally, regulation of cyclin B both at the transcriptional level and by its cytoplasmic localization, may also contribute to the G2/M arrest.23,24
Apoptotic Cell Death
In multicellular organisms, in addition to cell cycle arrest and repair, genotoxic stress can lead to the apoptotic demise of the damaged cell. DNA damage-induced cell deaths share the morphological characteristics of developmental programmed cell deaths, with features like cell rounding, cellular membrane blebbing, chromosomal condensation and DNA degradation shared between the two types of death.25
In mammals, two independent intracellular apoptotic signaling cascades can activate apoptosis: the mitochondrial pathway associated with activation of the apoptotic protease-activating factor-1 (Apaf-1) and caspase-9-containing apoptosome,26 and the death receptor pathway that acts through caspase-8.27 p53 contributes mainly to the activation of the former and exerts its function through the transcriptional regulation of its target genes. The mitochondrial pathway is regulated by the B-cell lymphoma 2 (Bcl-2) protein family, which includes both proapoptotic and prosurvival members. The sensors and mediators of apoptosis are the ‘BH3-only domain’ (BH3=BCL-2 homology domain 3) proapoptotic proteins. In response to DNA damage, at least two BH3 domain proteins, p53 upregulated modulator of apoptosis (PUMA) and Noxa, are transcriptionally induced in a p53-dependent manner.28,29 Post-translational phosphorylation (Bad, Bik) and proteolytic cleavage (Bid) are additional mechanisms for the regulation of the ‘BH3-only domain’ proteins in response to an apoptotic stimulus. The signal is then relayed to the ‘multidomain’ proapoptotic proteins Bax and Bak, which are initially kept at an inactive state. Upon activation, conformational changes result in their stable association into the mitochondrial membrane either to form a pore or interact with channel-forming proteins and increase membrane permeability. Cytochrome c and other intermembrane space proteins are then released to efficiently induce caspase-dependent and/or -independent apoptosis.30 In addition to PUMA and Noxa, a number of other p53 target genes, including but not limited to Bax, have been suggested to promote DNA damage - induced apoptosis.31
Programmed Cell Death in C. elegans
Significant progress in the field concerning the events in apoptosis was made from studies in the nematode C. elegans. In this species, two waves of apoptotic deaths have been found. The first occurs largely during embryogenesis, and helps to sculpt the cell lineages that produce all of the animal's somatic cells. The second wave of death occurs in the adult female germ line, where several hundred cells are eliminated during oogenesis.
Developmental cell death
During the somatic development of the animal, 131 of the 1090 cells generated undergo programmed cell death in a highly reproducible way: always the same cells die, and each cell dies at a characteristic point in development. Extensive genetic analysis of these cell deaths led to the identification of an evolutionarily conserved core apoptotic pathway that regulates all programmed cell deaths in C. elegans.32,33 Two genes, ced-3 and ced-4 (ced=cell death abnormal), are required for the killing process. The product of the former is a member of the caspase family of cysteine proteases; the product of the latter is homologous to mammalian Apaf-1 and functions genetically as a positive regulator of CED-3. A third gene, ced-9, protects cells that normally survive from undergoing programmed cell death. The CED-9 protein is homologous to the oncoprotein Bcl-2, which likewise promotes cell survival in mammals. Finally, EGL-1 is required for all developmental cell deaths in the animal. It belongs to a subset of Bcl-2 family members that contain only one of four Bcl-2 homology domains, the BH3 domain, and are thus known as BH3 domain only proteins. The BH3 domain allows EGL-1 to associate with and inhibit CED-9.
Biochemical studies have suggested that the key event required for apoptotic cell death in C. elegans is the processing of CED-3 from the inactive zymogen state into the active caspase.34,35 While the activation of CED-3 appears to be autocatalytic in nature, it does require association of the zymogen with oligomerized CED-4, likely forming a worm version of the apoptosome, which in mammals consists of at least three proteins – caspase-9, Apaf-1, and cytochrome c.36 In cells that should survive, formation of the apoptosome is prevented, at least in part because CED-4 is bound to and sequestered by CED-9 in a stable complex on the surface of mitochondria.37 Cells fated to die appear to be marked for apoptosis through the expression of EGL-1. Binding of EGL-1 to CED-9 causes the release of CED-4, which is then free to trigger the lethal proteolytic action of CED-3.38,39
Germ cell apoptosis
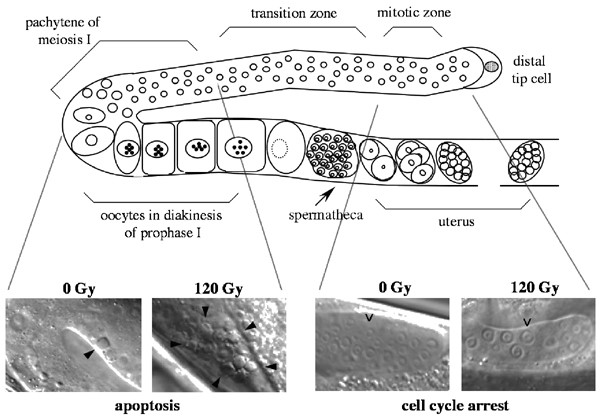
The C. elegans hermaphrodite consists of two U-shaped arms, joined proximally together at a common uterus (Figure 1). Unlike the somatic tissues, the germ line proliferates continuously both during larval development and in adult worms. In the adult hermaphrodite, germ cells progress through various stages of differentiation. The distal most germ cells proliferate mitotically and serve as a stem cell population. During their passage through a ‘transition zone’, they stop dividing and initiate meiosis. The most abundant population of cells resides in the pachytene stage of meiotic prophase that extends until before the bend of the gonad. Upon exit from this stage, they complete meiotic prophase, cellularize, undergo the final stages of maturation and finish meiosis after fertilization, which occurs as the oocyte pass through the spermatheca.40 Under normal growth conditions, approximately half of the female germ cells are doomed to die by programmed cell death.41 A steady-state level of zero to four apoptotic cells can be observed at any given time. These so-called ‘physiological germ cell deaths’ occur presumably to maintain tissue homeostasis.
Figure 1
DNA damage responses in the adult hermaphrodite C. elegans germ line. Mitotic stem cells proliferate throughout adulthood in the distal end of each gonad arm, then pass through a characteristic set of morphological stages as they undergo meiosis and descend towards the uterus. Following DNA damage germ cells in the mitotic region undergo proliferation arrest (right, open arrowheads), whereas meiotic germ cell nuclei undergo apoptosis (left, filled arrowheads)
DNA Damage Responses in C. elegans
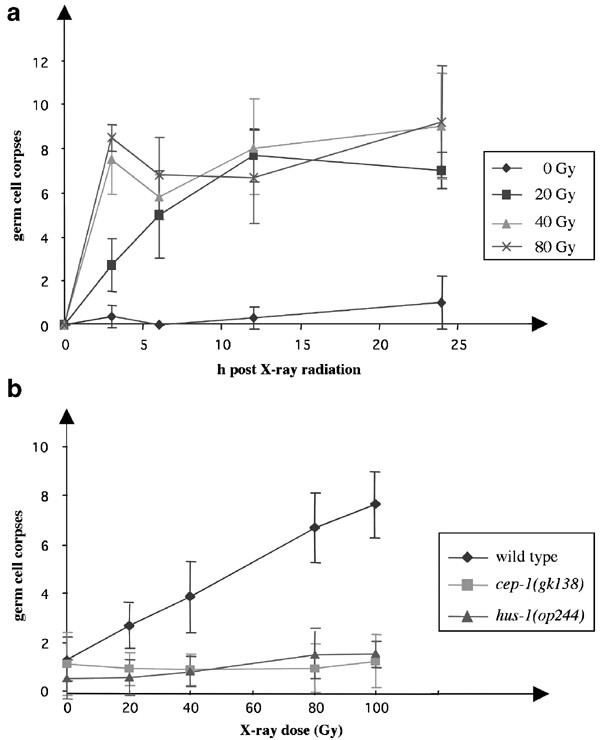
Studies in C. elegans on the effect of genotoxic stress have suggested that there are little or no checkpoint controls exerted during embryonic development, except for a possible transient intra-S checkpoint during early embryonic development.42 In the germ line, however, DNA damage-induced signaling induce two clear responses – cell cycle arrest and apoptosis – that are spatially separated (Figure 1). Exposure of worms to ionizing radiation causes a transient halt in cell cycle progression in the proliferating zone, resulting in a decreased number of mitotic germ cells. Notably, the volume of the arrested nuclei as well as that of the surrounding cytoplasm become enlarged, since cellular and nuclear growth continue to occur.43 In the meiotic compartment, after the exit from the pachytene region, increasing doses of ionizing radiation cause a dramatic increase in the number apoptotic cell deaths, which appear as early as 2–3 h after the insult, and persist for 20–60 min as highly refractile disks before being engulfed and degraded by the surrounding somatic sheath cells. Time course analyses have suggested that there are two waves of apoptosis, one early and one late, in response to irradiation (Figure 2a). The second wave of deaths might be caused by the delayed removal of cells that had been damaged in the mitotic region and failed to be repaired properly. Other types of DNA damage, such as treatment with the monofunctional alkylating agent ENU, or the accumulation of aberrant meiotic intermediates, also induce germ cell death, clearly showing that the deaths are a direct consequence of DNA damage.44
Figure 2
Wild-type worms respond with an early and a late wave of apoptotic deaths following exposure to ionizing radiation, in a dose-dependent manner (a). The checkpoint mutants cep-1 and hus-1 are defective in DNA damage-induced apoptotic cell death (b)
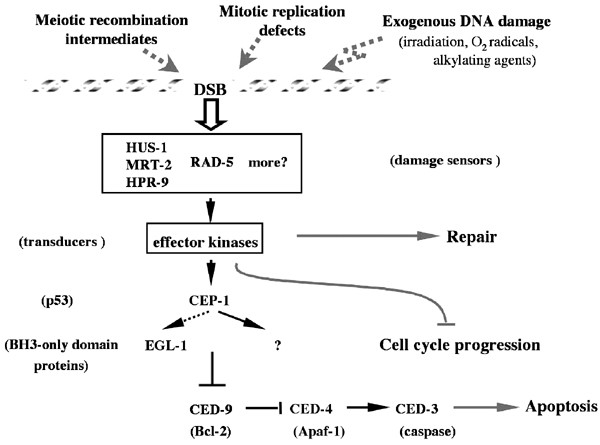
DNA damage-induced apoptosis requires the core apoptotic machinery (Figure 3): there is no cell death induced in the germ line of animals homozygous for the ced-3(n717) or ced-4(n1162) loss-of-function alleles. However, in contrast to physiological germ cell death, DNA damage-induced cell death is blocked by ced-9(n1950) gain-of-function and is severely reduced in the egl-1(n3082) (egl-1=egg-laying abnormal-1) loss-of-function mutation, indicating that the DNA damage response machinery is genetically distinct from the pathways that control somatic cell death and physiological germ cell death.
Figure 3
Exogenous DNA damage or damage in the form of meiotic recombination intermediates or mitotic replication defects is sensed by numerous proteins. HUS-1 and MRT-2, as part of a trimeric complex, and RAD-5 are involved in this process to allow for the action of the repair machinery. Cell cycle progression delay facilitates the repair process. The C. elegans p53 homolog, CEP-1, is responsible for the transcriptional activation of at least one BH3-only domain protein, EGL-1, that leads ultimately to the activation of the apoptotic machinery
Checkpoint Mutants in C. elegans
Various genetic screens for mutants defective in different signaling pathways, such as rad mutants and mutants with a high rate of chromosome nondisjunction, have revealed three strains defective for radiation-induced apoptosis: hus-1(op244), mrt-2(2663) (mrt=mortal germline-2) and rad-5(mn159) (Figure 3).43,45,46 All three mutations abrogate cell cycle arrest and apoptosis induced by DNA damage (Figure 2b), without affecting developmental or physiological germ cell death. Consistent with the situation in yeast and mammalian checkpoint gene mutants, they also show increased genomic instability and reduced long-term survival following genotoxic insults, as the high rates of embryonic lethality demonstrate.44,47
mrt-2 and hus-1 identify a conserved sensor pathway
Molecular characterization of mrt-2 and hus-1 showed that these genes encode the C. elegans homologs of S. pombe Rad1 and Hus1 (S. cerevisiae Rad17 and Mec3) checkpoint proteins.46,48 The fact that these two proteins are also required for DNA damage-induced cell cycle arrest in worms suggest that at least part of the DNA damage response pathway characterized in yeasts is conserved through evolution and also functions in nematodes. In fission yeast, Hus1, Rad1 and Rad9 form a heterotrimeric complex that resembles the PCNA complex in structure. Similarly, C. elegans HUS-1 is a nuclear protein that requires MRT-2 and the Rad9 homolog HPR-9 for proper localization to the nucleus. HUS-1 interacts physically with MRT-2, and a point mutation that disrupts this interaction compromises HUS-1 function.48 Furthermore, Hofmann et al48 recently showed that while HUS-1::GFP is uniformly associated with chromatin under normal conditions, upon DNA damage the fusion protein relocates within a few hours to distinct nuclear foci – possibly sites of unrepaired damage – consistent with the proposed function of the 9-1-1 complex as a marker of DNA damage.
Interestingly, while its role in cell cycle arrest appears to have been largely conserved from yeasts to nematodes, work in C. elegans has suggested that the 9-1-1 complex has acquired additional functions in nematodes that are either absent or not yet described in yeasts. Most surprise was the report by Ahmed and Hodgkin,46 that MRT-2 is required for telomere length maintenance: in mrt-2 mutants, telomeres progressively shorten over many generations, eventually leading to end-to-end chromosome fusion, genomic instability, and late-onset sterility by about the 15th generation. A similar defect has been described in hus-1 mutants.48 No such telomere defect is apparent in the yeast 9-1-1 mutants, suggesting either that this complex does not maintain telomere length in yeasts, or that it is redundant with other proteins in this function. Whether the 9-1-1 complex is also required for telomere length maintenance in mammals remains to be determined.
RAD-5/CLK-2 functions in parallel to the HUS-1/MRT-2 complex
A third checkpoint gene, rad-5, was originally identified in a screen for mutations that show reduced long-term survival following ionizing radiation. The mutation isolated in this screen, rad-5(mn159), was later found to be allelic to clk-2(qm37) (clk-2=clock gene), a mutation characterized by slow growth and a mild increase in animal life span.47 As with mrt-2 and hus-1, DNA damage-induced cell cycle arrest and apoptosis are abrogated in rad-5/clk-2 mutants. However, unlike MRT-2 and HPR-9, the C. elegans checkpoint protein RAD-5 is dispensable for the subcellular localization of HUS-1.48 Indeed, several lines of evidence suggest that RAD-5 functions in a pathway distinct from the 9-1-1 complex. First, long-term survival of hus-1; rad-5 and mrt-2; rad-5 double mutants are significantly reduced compared to the single mutants – this should not be the case if these proteins acted in a linear pathway. Second, unlike the 9-1-1 mutants, rad-5 mutants appear to also be defective in the S-phase replication checkpoint, similar to what is known for the yeast DNA polymerase epsilon.50 Third, rad-5 mutants do not show the mortal germ line phenotype of the 9-1-1 mutants. Finally, unlike the 9-1-1 complex, RAD-5 is an essential gene, as complete elimination of rad-5 gene function results in developmental arrest and embryonic lethality.
Surprisingly, mutations in TEL2, the budding yeast homolog of rad-5, do not result in any checkpoint defects or hypersensitivity to DNA damaging agents. Rather, hypomorphic tel2 mutants have been shown to bear short telomeres, suggesting a role for this gene in telomere maintenance.49 It is possible that the checkpoint role of RAD-5 is a new function acquired during metazoan evolution, and thus not present in yeasts. Alternatively, a role for Tel2p in DNA damage response might simply have been missed so far due to the essential nature of the gene.
Genes involved in meiotic DNA recombination
Double-strand breaks occur not only following genotoxic stress, but also normally during meiotic prophase to initiate meiotic recombination events.51 Two coupled processes, the formation and the processing of DSBs are involved in meiotic recombination. The SPO11 gene product is responsible for enzymatic DNA cleavage to create DSBs and Mre11 subsequently processes these through its intrinsic exonuclease activity. Rad51, a member of the RecA-strand exchange protein family, catalyzes the invasion of the single-strand DNA overhangs generated by Mre11 into a recipient homologous double-strand DNA molecule, thereby initiating the formation of D loops and the later steps of meiotic recombination.51,52 Mutations in Rad51 confer an enhanced sensitivity to DNA damaging agents in yeast, a reduction in mitotic recombination and impaired meiosis. Silencing of the gene via RNAi in C. elegans results in high levels of embryonic lethality and increased frequency of males,44,53,54 phenotypes encountered commonly in meiotic mutants. Moreover, a dramatic increase in germ cell apoptosis is observed when rad-51 is inactivated. When meiotic recombination is blocked by the absence of sporulation, meiosis-specific protein (SPO-11), this increase no longer occurs, suggesting that the resulting deaths are indeed triggered by accumulation of recombination intermediates, which are perceived as a form of DNA damage. Mutations in hus-1, mrt-2 and rad-5 checkpoint genes suppress the _rad-51_-dependent apoptotic death, suggesting that disruption of the DNA damage checkpoint pathway compromises the ability of the cells to sense and respond to the incurred damage.44 Damage caused by radiation treatment in late embryonic stages, in animals depleted from RAD-51, resulted in several developmental defects in vulva and gonad formation. This effect is likely due to inability of repair of radiation-induced DSBs, and implies that rad-51 also functions in DNA damage response in the soma.55
As is the case with rad-51, loss of mre-11 function results in defective meiotic recombination and increased germ cell apoptosis. Consistent with this, the viability of progeny in these mutants drops dramatically. In mammals, Mre11 has been implicated in DNA damage response.56 However, the fact that mre-11 mutants show increased, rather than decreased germ cell apoptosis argues that in C. elegans, the DNA damage checkpoint is still functional in mre-11 mutants.57
Another crucial component of the C. elegans meiotic recombination machinery is a germline-specific member of the MutS protein family, MSH-5.58 Crossing over and chiasma formation events depend on msh-5 gene (_msh_=mismatch repair gene) function: in msh-5 mutants, both events are severely reduced or eliminated. Nevertheless, C. elegans msh-5 mutants are able to complete meiosis and gametogenesis with high efficiency and produce embryos without undergoing germ cell apoptosis like their murine counterparts.59,60 It is likely that msh-5 mutant cells escape apoptosis because they are still proficient in repairing dsDNA breaks, but in a way that does not lead to crossovers between homologous chromosomes.58
Mismatch repair genes
Unlike Msh5 and its heterodimer partner Msh4, which are involved in the control of meiotic crossing over, the other members of the MutS protein family have a substantial role in the correction of DNA polymerase errors, through mechanisms such as postreplication DNA mismatch repair (MMR). In yeast, the Msh2 protein participates in all three different complexes to repair base–base mismatches and DNA loops61 and mutations in the human and mouse gene result in tumor development.62 The tumorigenic potential of MMR mutants is likely due to an increased mutation rate in these cells, coupled with a resistance to apoptosis caused by certain mutagens, which would allow the mutated cells to survive.62
In C. elegans, msh-2 mutants also show a mutator phenotype, as well as reduced fertility and long-term viability (possibly due to the accumulation of detrimental mutations). msh-2 mutants also show increased microsatellite instability. Following exposure to ionizing radiation, the apoptotic response of msh-2 mutants is impaired and/or delayed, but not abrogated. It is possible that binding of the lesions by mismatch repair proteins promotes both the DNA repair process and the elimination of cells by apoptosis, depending on the amount of damage.63 The role of MSH-2 in apoptosis, though, still remains to be clarified.
No role for CHK-2 in DNA damage response in C. elegans?
Unlike the situation in other organisms, where Chk2 proteins participate in several distinct signaling pathways that maintain genome integrity, C. elegans chk-2 mutants show normal checkpoint responses following DNA damage. While it is possible that in C. elegans, CHK-2 is simply redundant with another protein for its well-characterized checkpoint function, an alternative explanation is that in nematodes, CHK-2 has simply lost its role in DNA damage response and has acquired a different function. Indeed, elegant genetic studies have suggested that C. elegans CHK-2 is required for establishment of homolog alignment and synapsis.64 An early role in the initiation of meiotic recombination was also suggested by the observation that chk-2 mutations, like those in the meiotic endonuclease gene spo-11, suppress the increase in apoptosis normally observed in worms depleted of RAD-51. Alternatively, chk-2 mutants might be successful in initiating recombination, but defective in activating the checkpoint in response to a subset of DNA lesions, such as recombination intermediates.
The C. elegans p53 homolog CEP-1
The tumor suppressor protein p53 plays a key role in the integration of cellular responses to genotoxic stimuli. In higher organisms, it acts as a guardian of genome integrity, by inducing cell cycle arrest or apoptosis following DNA damage. p53-mediated apoptosis is a consequence of the combined expression of various target-genes, either through its transactivation function or transcriptional repression.65,66 Like its mammalian homolog, the C. elegans p53 protein, CEP-1, is required for DNA damage-induced germ cell apoptosis (Figure 2b).67,68 In contrast to mammalian p53, however, CEP-1 is dispensable for cell cycle arrest activation, a property shared also by Drosophila p53.69 It is likely that this primordial proapoptotic function depends on transcriptional activation of genes that act on the core apoptotic machinery.
Indeed, recent studies have suggested that induction of germ cell apoptosis following DNA damage is transcriptionally regulated: following DNA damage, mRNA levels for the BH3 domain protein EGL-1 are dramatically upregulated, consistent with the previously described role for egl-1 in DNA damage induced apoptosis in the adult germ line. Interestingly, egl-1 mRNA levels do not change following DNA damage in cep-1 mutants, nor in any of the other checkpoint mutants thought to act upstream of cep-1 (hus-1, rad-5). An attractive model consistent with these observations would be that CEP-1 acts as a direct transactivator for the egl-1 gene following DNA damage (Figure 3). A particular appeal of this model is its similarity with the situation in mammals, where DNA damage-induced apoptosis requires the p53-dependent transcriptional activation of the BH3 domain proteins Noxa and PUMA. How CEP-1 becomes activated following DNA damage, and whether it can directly bind to the egl-1 regulatory region remains to be determined.
Conclusions
Loss of DNA damage checkpoint function can lead to genome instability, one of the driving forces towards carcinogenesis. Understanding how checkpoint pathways maintain genome integrity is thus crucial for a complete understanding of the origin of cancer and of other genetic abnormalities. Such knowledge might ultimately provide insights into novel ways to prevent, diagnose, and treat such diseases. Considering the fact that DNA damage checkpoints in higher eukaryotes are more complicated than those found in the unicellular yeasts, an easily accessible metazoan model organism is required to study these biological processes in depth. C. elegans is a promising candidate for this function. Its powerful genetics and molecular tools allow an in-depth study of developmental and various other biological pathways. Indeed, the advances made so far in the area of DNA damage response suggest that this species can provide a useful jumping board, from which acquired knowledge can be used to test specific hypotheses in the human system. As in many other biological problems, the worm's simplicity may be the key to unveiling the complexities of the human organism.
Abbreviations
Rad:
radiation sensitive
PCNA:
proliferating Cell Nuclear Antigen
Hus1:
hydroxyurea-sensitive 1
Mre11:
meiotic recombination 11
Nbs1:
nibrin (mutated in Nijmegen breakage syndrome)
BRCA1:
breast cancer 1
ATM:
ataxia telangiectasia mutated kinase
ATR:
ataxia telangiectasia-related kinase
Tel1:
telomere length regulation 1
Chk1:
checkpoint kinase 1
Cdk:
cyclin-dependent kinase
Cdc:
cell division control
Apaf-1:
apoptotic protease-activating factor-1
Bcl-2:
B-cell lymphoma 2
BH3:
BCL-2 homology domain 3
PUMA:
p53 upregulated modulator of apoptosis
ced:
cell death abnormal
egl-1 :
egg-laying abnormal-1
mrt-2 :
mortal germ line-2
clk-2 :
clock gene
Spo 11:
sporulation, meiosis-specific protein
msh :
mismatch repair gene
cep-1 :
C. elegans p53
References
- Griffiths DJ, Barbet NC, McCready S, Lehmann AR and Carr AM (1995) Fission yeast rad17: a homologue of budding yeast RAD24 that shares regions of sequence similarity with DNA polymerase accessory proteins. EMBO J. 14: 5812–5823
Article CAS PubMed PubMed Central Google Scholar - Green CM, Erdjument-Bromage H, Tempst P and Lowndes NF (2000) A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr. Biol. 10: 39–42
Article CAS PubMed Google Scholar - Tsurimoto T and Stillman B (1991) Replication factors required for SV40 DNA replication in vitro. II. Switching of DNA polymerase alpha and delta during initiation of leading and lagging strand synthesis. J. Biol. Chem. 266: 1961–1968
CAS PubMed Google Scholar - Zou L, Cortez D and Elledge SJ (2002) Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 16: 198–208
Article CAS PubMed PubMed Central Google Scholar - Chamankhah M and Xiao W (1999) Formation of the yeast Mre11–Rad50–Xrs2 complex is correlated with DNA repair and telomere maintenance. Nucleic Acids Res. 27: 2072–2079
Article CAS PubMed PubMed Central Google Scholar - Maser RS, Monsen KJ, Nelms BE and Petrini JH (1997) hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol. Cell Biol. 17: 6087–6096
Article CAS PubMed PubMed Central Google Scholar - Shiloh Y (1997) Ataxia-telangiectasia and the Nijmegen breakage syndrome: related disorders but genes apart. Annu. Rev. Genet. 31: 635–662
Article CAS PubMed Google Scholar - Gowen LC, Avrutskaya AV, Latour AM, Koller BH and Leadon SA (1998) BRCA1 required for transcription-coupled repair of oxidative DNA damage. Science 281: 1009–1012
Article CAS PubMed Google Scholar - Moynahan ME, Chiu JW, Koller BH and Jasin M (1999) Brca1 controls homology-directed DNA repair. Mol. Cell 4: 511–518
Article CAS PubMed Google Scholar - Snouwaert JN, Gowen LC, Latour AM, Mohn AR, Xiao A, DiBiase L and Koller BH (1999) BRCA1 deficient embryonic stem cells display a decreased homologous recombination frequency and an increased frequency of non-homologous recombination that is corrected by expression of a brca1 transgene. Oncogene 18: 7900–7907
Article CAS PubMed Google Scholar - Hartwell LH and Kastan MB (1994) Cell cycle control and cancer. Science 266: 1821–1828
Article CAS PubMed Google Scholar - Zhou BB and Elledge SJ (2000) The DNA damage response: putting checkpoints in perspective. Nature 408: 433–439
Article CAS PubMed Google Scholar - Khanna KK and Jackson SP (2001) DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 27: 247–254
Article CAS PubMed Google Scholar - Shiloh Y (2001) ATM and ATR: networking cellular responses to DNA damage. Curr. Opin. Genet. Dev. 11: 71–77
Article CAS PubMed Google Scholar - Walworth NC and Bernards R (1996) rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science 271: 353–356
Article CAS PubMed Google Scholar - Gerald JN, Benjamin JM and Kron SJ (2002) Robust G1 checkpoint arrest in budding yeast: dependence on DNA damage signaling and repair. J. Cell Sci. 115: 1749–1757
CAS PubMed Google Scholar - Ekholm SV and Reed SI (2000) Regulation of G(1) cyclin-dependent kinases in the mammalian cell cycle. Curr. Opin. Cell Biol. 12: 676–684
Article CAS PubMed Google Scholar - Sherr CJ and Roberts JM (1999) CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13: 1501–1512
Article CAS PubMed Google Scholar - Bartek J and Lukas J (2001) Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr. Opin. Cell Biol. 13: 738–747
Article CAS PubMed Google Scholar - Graves PR, Lovly CM, Uy GL and Piwnica-Worms H (2001) Localization of human Cdc25C is regulated both by nuclear export and 14-3-3 protein binding. Oncogene 20: 1839–1851
Article CAS PubMed Google Scholar - Matsuoka S, Huang M and Elledge SJ (1998) Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science 282: 1893–1897
Article CAS PubMed Google Scholar - Peng CY, Graves PR, Thoma RS, Wu Z, Shaw AS and Piwnica-Worms H (1997) Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science 277: 1501–1505
Article CAS PubMed Google Scholar - Jin P, Hardy S and Morgan DO (1998) Nuclear localization of cyclin B1 controls mitotic entry after DNA damage. J. Cell Biol. 141: 875–885
Article CAS PubMed PubMed Central Google Scholar - Toyoshima F, Moriguchi T, Wada A, Fukuda M and Nishida E (1998) Nuclear export of cyclin B1 and its possible role in the DNA damage-induced G2 checkpoint. EMBO J. 17: 2728–2735
Article CAS PubMed PubMed Central Google Scholar - Horvitz HR, Ellis HM and Sternberg PW (1982) Programmed cell death in nematode development. Neurosci. Comment. 1: 56–65
Google Scholar - Green DR and Reed JC (1998) Mitochondria and apoptosis. Science 281: 1309–1312
Article CAS PubMed Google Scholar - Ashkenazi A and Dixit VM (1998) Death receptors: signaling and modulation. Science 281: 1305–1308
Article CAS PubMed Google Scholar - Nakano K and Vousden KH (2001) PUMA, a novel proapoptotic gene, is induced by p53. Mol. Cell 7: 683–694
Article CAS PubMed Google Scholar - Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, Tokino T, Taniguchi T and Tanaka N (2000) Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 288: 1053–1058
Article CAS PubMed Google Scholar - Borner C (2003) The Bcl-2 protein family: sensors and checkpoints for life-or-death decisions. Mol. Immunol. 39: 615–647
Article CAS PubMed Google Scholar - Miyashita T and Reed J (1995) Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 80: 293–299
Article CAS PubMed Google Scholar - Hengartner MO and Horvitz HR (1994a) The ins and outs of programmed cell death during C. elegans development. Philos. Trans. R. Soc. Lond. B Biol. Sci. 345: 243–246
Article CAS PubMed Google Scholar - Hengartner MO and Horvitz HR (1994b) Programmed cell death in Caenorhabditis elegans. Curr. Opin. Genet. Dev. 4: 581–586
Article CAS PubMed Google Scholar - Xue D, Shaham S and Horvitz HR (1996) The Caenorhabditis elegans cell-death protein CED-3 is a cysteine protease with substrate specificities similar to those of the human CPP32 protease. Genes Dev. 10: 1073–1083
Article CAS PubMed Google Scholar - Chinnaiyan AM, O'Rourke K, Lane BR and Dixit VM (1997) Interaction of CED-4 with CED-3 and CED-9: a molecular framework for cell death. Science 275: 1122–1126
Article CAS PubMed Google Scholar - Yang X, Chang HY and Baltimore D (1998) Essential role of CED-4 oligomerization in CED-3 activation and apoptosis. Science 281: 1355–1357
Article CAS PubMed Google Scholar - Chen F, Hersh BM, Conradt B, Zhou Z, Riemer D, Gruenbaum Y and Horvitz HR (2000) Translocation of C. elegans CED-4 to nuclear membranes during programmed cell death. Science 287: 1485–1489
Article CAS PubMed Google Scholar - Conradt B and Horvitz HR (1998) The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell 93: 519–529
Article CAS PubMed Google Scholar - del Peso L, Gonzalez VM, Inohara N, Ellis RE and Nunez G (2000) Disruption of the CED-9.CED-4 complex by EGL-1 is a critical step for programmed cell death in Caenorhabditis elegans. J. Biol. Chem. 275: 27205–27211
CAS PubMed Google Scholar - Schedl T (1997) Developmental genetics of the germ line. In C. elegans II, Riddle DL, Blumenthal T, Meyer BJ and Priess JR (eds) (Plainview, NY: Cold Spring Harbor Laboratory Press), pp. 241–269
- Gumienny TL, Lambie E, Hartwieg E, Horvitz HR and Hengartner MO (1999) Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development 126: 1011–1022
CAS PubMed Google Scholar - Encalada SE, Martin PR, Phillips JB, Lyczak R, Hamill DR, Swan KA and Bowerman B (2000) DNA replication defects delay cell division and disrupt cell polarity in early Caenorhabditis elegans embryos. Dev. Biol. 228: 225–238
Article CAS PubMed Google Scholar - Hodgkin J, Horvitz HR and Brenner S (1979) Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics 91: 67–94
CAS PubMed PubMed Central Google Scholar - Gartner A, Milstein S, Ahmed S, Hodgkin J and Hengartner MO (2000) A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Mol. Cell 5: 435–443
Article CAS PubMed Google Scholar - Hartman PS and Herman RK (1982) Radiation-sensitive mutants of Caenorhabditis elegans. Genetics 102: 159–178
CAS PubMed PubMed Central Google Scholar - Ahmed S and Hodgkin J (2000) MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature 403: 159–164
Article CAS PubMed Google Scholar - Ahmed S, Alpi A, Hengartner MO and Gartner A (2001) C. elegans RAD-5/CLK-2 defines a new DNA damage checkpoint protein. Curr. Biol. 11: 1934–1944
Article CAS PubMed Google Scholar - Hofmann ER, Milstein S, Boulton SJ, Ye M, Hofmann JJ, Stergiou L, Gartner A, Vidal M and Hengartner MO (2002) Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Curr. Biol. 12: 1908–1918
Article CAS PubMed Google Scholar - Lustig AJ and Petes TD (1986) Identification of yeast mutants with altered telomere structure. Proc. Natl. Acad. Sci. USA 83: 1398–1402
Article CAS PubMed PubMed Central Google Scholar - Navas TA, Zhou Z and Elledge SJ (1995) DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell 80: 29–39
Article CAS PubMed Google Scholar - Roeder GS (1997) Meiotic chromosomes: it takes two to tango. Genes Dev. 11: 2600–2621
Article CAS PubMed Google Scholar - Bishop DK, Park D, Xu L and Kleckner N (1992) DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69: 439–456
Article CAS PubMed Google Scholar - Takanami T, Mori A, Takahashi H and Higashitani A (2000) Hyper-resistance of meiotic cells to radiation due to a strong expression of a single recA-like gene in Caenorhabditis elegans. Nucleic Acids Res. 28: 4232–4236
Article CAS PubMed PubMed Central Google Scholar - Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M and Villeneuve AM (1998) Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94: 387–398
Article CAS PubMed Google Scholar - Rinaldo C, Bazzicalupo P, Ederle S, Hilliard M and La Volpe A (2001) Roles for Caenorhabditis elegans rad-51 in meiosis and in resistance to ionizing radiation during development. Genetics 160: 471–479
Google Scholar - D'Amours D and Jackson SP (2002) The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 3: 317–327
Article CAS PubMed Google Scholar - Chin GM and Villeneuve AM (2001) C. elegans mre-11 is required for meiotic recombination and DNA repair but is dispensable for the meiotic G(2) DNA damage checkpoint. Genes Dev. 15: 522–534
Article CAS PubMed PubMed Central Google Scholar - Kelly KO, Dernburg AF, Stanfield GM and Villeneuve AM (2000) Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics 156: 617–630
CAS PubMed PubMed Central Google Scholar - de Vries SS, Baart EB, Dekker M, Siezen A, de Rooij DG, de Boer P and te Riele H (1999) Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis. Genes Dev. 13: 523–531
Article CAS PubMed PubMed Central Google Scholar - Edelmann W, Cohen PE, Kneitz B, Winand N, Lia M, Heyer J, Kolodner R, Pollard JW and Kucherlapati R (1999) Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat. Genet. 21: 123–127
Article CAS PubMed Google Scholar - Harfe BD and Jinks-Robertson S (2000) DNA mismatch repair and genetic instability. Annu. Rev. Genet. 34: 359–399
Article CAS PubMed Google Scholar - Buermeyer AB, Deschenes SM, Baker SM and Liskay RM (1999) Mammalian DNA mismatch repair. Annu. Rev. Genet. 33: 359–399
Article Google Scholar - Degtyareva NP, Greenwell P, Hofmann ER, Hengartner MO, Zhang L, Culotti JG and Petes TD (2002) Caenorhabditis elegans DNA mismatch repair gene msh-2 is required for microsatellite stability and maintenance of genome integrity. Proc. Natl. Acad. Sci. USA 99: 2158–2163
Article CAS PubMed PubMed Central Google Scholar - MacQueen AJ and Villeneuve AM (2001) Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Genes Dev. 15: 1674–1687
Article CAS PubMed PubMed Central Google Scholar - Kastan MB, Zhan Q, el-Deiry WS, Carrier F, Jacks T, Walsh WV, Plunkett BS, Vogelstein B and Fornace Jr AJ (1992) A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell 71: 587–597
Article CAS PubMed Google Scholar - Moberg KH, Tyndall WA and Hall DJ (1992) Wild-type murine p53 represses transcription from the murine c-myc promoter in a human glial cell line. J. Cell Biochem. 49: 208–215
Article CAS PubMed Google Scholar - Derry WB, Putzke AP and Rothman JH (2001) Caenorhabditis elegans p53: role in apoptosis, meiosis, and stress resistance. Science 294: 591–595
Article CAS PubMed Google Scholar - Schumacher B, Hofmann K, Boulton S and Gartner A (2001) The C. elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis. Curr. Biol. 11: 1722–1727
Article CAS PubMed Google Scholar - Ollmann M, Young LM, Di Como CJ, Karim F, Belvin M, Robertson S, Whittaker K, Demsky M, Fisher WW, Buchman A, Duyk G, Friedman L, Prives C and Kopczynski C (2000) Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell 101: 91–101
Article CAS PubMed Google Scholar
Author information
Authors and Affiliations
- Institute of Molecular Biology, University of Zurich, Winterthurerstrasse 190, Zurich, CH-8057, Switzerland
L Stergiou & M O Hengartner
Authors
- L Stergiou
You can also search for this author inPubMed Google Scholar - M O Hengartner
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toM O Hengartner.
Additional information
Edited by G Melino
Rights and permissions
About this article
Cite this article
Stergiou, L., Hengartner, M. Death and more: DNA damage response pathways in the nematode C. elegans.Cell Death Differ 11, 21–28 (2004). https://doi.org/10.1038/sj.cdd.4401340
- Received: 30 June 2003
- Accepted: 05 August 2003
- Published: 19 December 2003
- Issue Date: 01 January 2004
- DOI: https://doi.org/10.1038/sj.cdd.4401340