Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast (original) (raw)
Introduction
Macromolecule metabolism in cells constantly needs to be finely tuned depending on environmental nutritional conditions. Nutrient limitation in yeast cells1 and/or hormonal stimulation in mammalian cells2 trigger a conserved catabolic ‘self-eating’ process called autophagy, which is responsible for the degradation and recycling of nonessential cellular components (from macromolecules to whole organelles). Autophagy allows cells to survive extended periods of starvation, and thus lengthens their lifespan. However, the completion of this ‘self-eating process’ will ultimately lead to cell death and hence, autophagy is also envisaged in higher eukaryotes as a programmed cell death (PCD).3
Central to this programme that controls the balance between biosynthesis and degradation is Target Of Rapamycin (TOR), a phosphatidylinositol kinase-related protein kinase that is conserved from yeast to mammals. In response to nutrients, TOR controls cell growth through the regulation of translation, ribosome biosynthesis, and transcription of a subset of mRNAs. TOR is active in the presence of nutrients and inactive upon nutrient limitation. Starvation-induced inhibition of TOR can be mimicked by the presence of rapamycin (Rapa), which blocks the cell cycle in early G1 phase, driving cells into a G0 state, and eventually triggering autophagy.4
Based on morphological characteristics, two types of autophagy are defined. The best documented is macroautophagy. Most of the mechanistic insights come from genetic studies carried out in yeast Saccharomyces cerevisiae. Some similar, but not identical, mammalian counterparts of the yeast autophagic machinery have been identified, and a unified nomenclature was recently adopted.5 Macroautophagy consists of a random sequestration of cytosolic portions in double-membrane-bound vesicles called autophagosomes.1 Vesicular factors mediate the fusion of the outer membrane of autophagosomes with autolysosomes in mammals or with vacuoles in yeast.6, 7 The resulting single-membrane-bound autophagic bodies released into autolysosomes or vacuoles8 are further degraded by lysosomal/vacuolar soluble acid hydrolases.
The second form of autophagy, called microautophagy is less characterised but is also found in higher eukaryotes.9 Most of the mechanistic perception comes from genetic studies carried out in methylotrophic yeast Pichia pastoris,10 in which metabolic adaptation is caused by changes in carbon source that drives pexophagy (degradation of peroxisomes) by either macro or microautophagy.11 Morphologically, the hallmarks of microautophagy are indentations in the vacuolar or lysosomal boundary membrane that allow direct uptake of the cytosol and/or organelles. Pinching off of these invaginations releases microautophagic vesicles into the vacuolar lumen where they will be degraded. Diverging descriptions of microautophagy have been documented in S. cerevisiae, concerning its dependence on the Apg/Aut pathway12 and TOR.13
In higher eukaryotes, complete autophagy is generally acknowledged as a PCD. Recent studies report that type II PCD (autophagy) could substitute for defective type I PCD (apoptosis), suggesting (i) that autophagy could be physiologically triggered by other means besides nutritional stress, and (ii) that autophagy could be an alternative to apoptosis.14, 15 It has therefore become essential to characterise the interactions (if any) between type I and type II PCD. During apoptosis, mitochondria are acknowledged as a central element; recently, some reports have suggested the involvement of mitochondria in autophagy also.16, 17, 18, 19
Here, we report for the first time, the occurrence of autophagy as a consequence of a physiological mitochondrial dysfunction in yeast, resulting from either anaerobic and heat stress growth of a FMC1 null mutant or anaerobic growth of a strain carrying a point mutation in the ATP2 gene. This mitochondrial dysfunction results in the collapse of the electrical potential across the mitochondrial inner membrane (ΔΨ), while not decreasing the cellular ATP concentration. This newly described induction of autophagy results in the preferential removal of mitochondria, and ultimately leads to cell death. Mitochondrial damage-induced autophagy is therefore discussed as a possible cellular survey over the quality control of mitochondria or as an ultimate programme initiated when cells cannot recover from mitochondrial injury.
Results
Anaerobic shift of FMC1 null mutant at 37°C induces autophagy
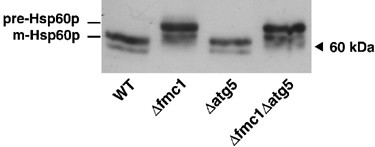
Catalytic core of mitochondrial ATP synthase (F1 sector) is composed of five different proteins arranged as follows: α_3_β_3_γδɛ. The correct assembly of _α_3–_β_3 hexamer into a functional F1 sector requires molecular chaperones Atp12p and Atp11p. At elevated temperatures, Atp12p stability seems to be challenged in the absence of Fmc1p, a nuclear-encoded soluble protein located in the mitochondrial matrix. Therefore, Fmc1p is also required, although only under heat stress conditions (37°C), for the proper assembly of _α_-F1 and β_-F1 subunits of mitochondrial ATP synthase.20 Cells lacking Atp12p or Atp11p, or cells lacking Fmc1p and grown at restrictive temperature, accumulate large electron-dense particles in the mitochondrial matrix, which were shown by ultrastructural and immunocytochemical analysis to be composed almost exclusively of mature F1 α and/or β subunits (i.e. processed to remove the amino-terminal targeting peptide).20, 21, 22 These mutants hence lack functional ATP synthase. Such a defect becomes a major issue in respiration-deficient cells, where the maintenance of an electrical potential across the mitochondrial inner membrane (ΔΨ) is no longer provided by the respiratory chain, but should be generated by the reverse functioning of the mitochondrial ATP synthase and the mitochondrial ATP/ADP carrier (ANC): normally in nonrespiring cells, the ANC imports ATP and exports ADP, and the F1-catalysed ATP hydrolysis is coupled to proton extrusion toward the mitochondrial inter-membrane space, thus maintaining a weaker ΔΨ. Hence, respiration-deficient cells harbouring a defective F1 sector are unable per se to maintain the ΔΨ required for the importation of mitochondrial proteins and therefore exhibit a general defect in mitochondrial biogenesis.23, 24 The severe alteration in mitochondrial protein import of anaerobically grown Δ_fmc1 mutants (at 37°C) provides indirect evidence of such a ΔΨ collapse. Indeed, Western blot analysis (Figure 1) of total protein extracts showed an accumulation of the precursor form of the nuclear-encoded mitochondrial Hsp60p (pre-Hsp60p), whereas only the mature form (m-Hsp60p) is detected in the wild-type (WT) cells, thus confirming that mitochondrial ΔΨ is impaired in our model, in agreement with the previously published data.25
Figure 1
Alterations in the processing of nuclear-encoded mitochondrial HSP60p. Cells were grown anaerobically for 24 h at 37°C, and 30 μ_g total protein extracts were analysed on a 9–15% gradient SDS-PAGE. Immunodetection was performed with anti-HSP60 antibody. Pre-Hsp60p: precursor form; m-Hsp60p: mature form. Wild-type (WT), strain MCP1-8; Δ_fmc1 cells, strain MCP6-8; Δ_atg5_ cells, strain MCP185; Δ_fmc1_Δ_atg5_ cells, strain MCP685. Western blot is representative of at least five independent experiments, all supporting the same conclusion
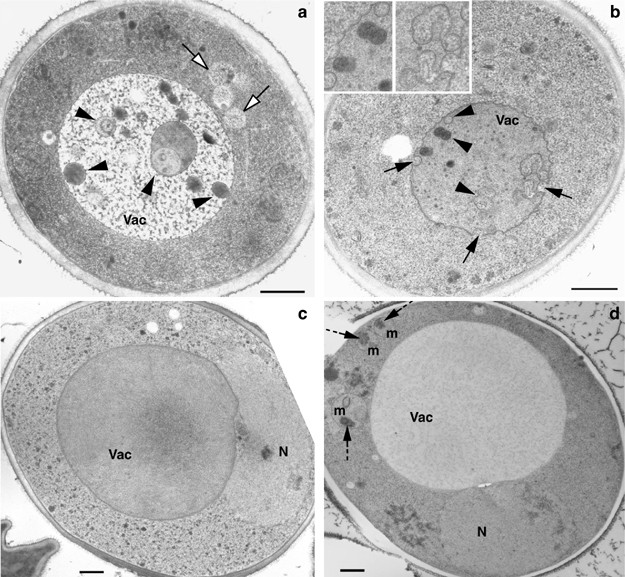
Electron micrographs (EM) of a Δ_fmc1_ mutant grown under nonstarving conditions at 37°C and under anaerobiosis (−O2) revealed the occurrence of a process morphologically similar to autophagy (Figure 2a and b), whereas WT cells under similar conditions (37°C −O2 for 24 h: Figure 2c, or for 48 h: Supplementary Data) or Δ_fmc1_ cells grown in the presence of O2 at 37°C (Figure 2d) or −O2 at 28°C (not shown) appeared normal. As soon as 24 h after an anaerobic shift, some Δ_fmc1_ cells exhibited macroautophagic phenotypes (Figure 2a) including cytosolic autophagosomes (white arrows) and autophagic bodies in vacuoles (arrowheads). Other cells showed characteristics similar to microautophagy (Figure 2b), namely vacuolar indentations (black arrows), and autophagic bodies sequestered within the vacuole.
Figure 2
EM analysis of Δ_fmc1_ mutant and WT strain grown with or without oxygen at 37°C. Cells were pregrown under aerobic conditions in YPD at 37°C for 12 h, and then shifted (a–c) or not (d) to anaerobiosis at 37°C. (a) EM of Δ_fmc1_ cells (strain MC6) 48 h after anaerobiosis: cell showing typical macroautophagic features with autophagosomes appearing in the cytosol (white arrows) and autophagic bodies in the vacuole (black arrowheads) (b) EM of Δ_fmc1_ cells (strain MC6) 24 h after anaerobiosis: cells display typical features of microautophagy with deep indentations of the vacuolar boundary membrane (arrows) and autophagic bodies sequestered within (arrowheads). (c) EM of WT cells (strain MC1) 24 h after anaerobiosis at 37°C. (d) EM of Δ_fmc1_ cells (strain MC6) grown under aerobic conditions at 37°C for 48 h. Mitochondria exhibit protein aggregates in their matrix (dashed arrows). Vac, vacuole; N, nucleus; m, mitochondria. Scale bar 0.5 _μ_m
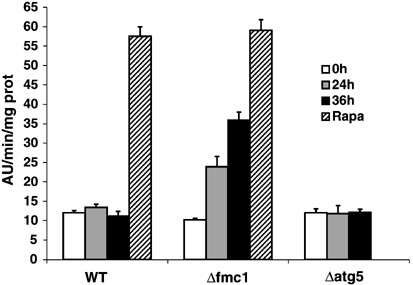
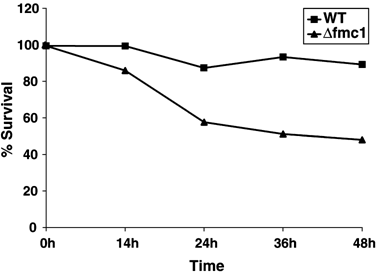
In order to confirm that these phenotypes were relevant to an autophagic process, we examined the autophagic activity in Δ_fmc1_ strain versus WT or autophagy-deficient Δ_atg5_ strain. ATG5 encodes an essential component of the macroautophagy machinery required for the formation/completion of autophagosomes, and its disruption results in defective macroautophagy.26, 27 Autophagic activity was assayed using the alkaline phosphatase (ALP) reporter test28 (Figure 3) in which the delivery of cytosolic N-terminal-truncated Pho8Δ60p in the vacuole results from its random trapping in autophagic vesicles; the subsequent vacuolar Pep4p-mediated activation allows the quantification of the process. Autophagic activity in WT was constant with time and remained at background levels, which was probably due to the participation in the reaction of Pho13p, another alkaline phosphatase that is constitutively active. However, this background activity was independent of autophagy as it was comparable to the level observed in the Δ_atg5_ strain. In Δ_fmc1_ cells, however, ALP activity was found to increase over time: after 24 h −O2 (grey bars), the activity was significantly increased as compared to t_=0 h or to WT and this difference was further enhanced after 36 h (black bars), reaching 62% of the maximal autophagic activity determined by 24 h-Rapa treatment (hatched bars). Longer anaerobic treatment (48 h) of Δ_fmc1 cells resulted in the collapse of ALP activity, and this decrease was found to correlate with more than 50% cell death (Figure 4).
Figure 3
ALP assays. Cells were pregrown under aerobic conditions in YPD for 12 h at 37°C. Supernatants of permeabilised cells were assayed for autophagic activity before anaerobic shift (white bars), or 24 h (grey bars) and 36 h (black bars) after anaerobiosis. Anaerobically grown cells were treated with rapamycin for 24 h and ALP activity assayed (hatched bars). The activities presented with the SEM are the average of three independent experiments. WT, strain MCP1-8; Δ_fmc_1 cells, strain MCP6-8; Δ_atg5_ cells, strain MCP185
Figure 4
Survival after anaerobic growth. Wild-type (squares) and Δ_fmc1_ cells (triangles) were adapted overnight at 37°C under aerobiosis. At 0 h, cells were shifted to 37°C, −O2. At the indicated times, aliquots were taken and 200 cells were plated on YPD plates and returned to permissive temperature. Colonies were counted 3 days after plating. Results are expressed as a percentage of colonies counted before anaerobic shift, and are representative of three independent experiments
Taken together, these biochemical data confirmed the EM observations, namely that an anaerobic shift of Δ_fmc1_ mutant to 37°C triggered autophagy independently of any nutritional stress, but as a consequence of a mitochondrial defect. The data also suggest that cell death might accompany autophagy.
Autophagy driven by mitochondrial defects triggers preferential degradation of mitochondria and is dependent on the ATG machinery
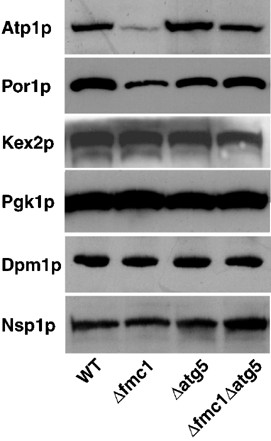
In order to assay whether autophagy triggered by mitochondrial defects could target damaged mitochondria, we examined the amount of several proteins in Δ_fmc1_ cells compared to WT, Δ_atg5_, and Δ_fmc1_ Δ_atg5_ cells, all grown under autophagy-inducing conditions (−O2, 37°C) for 24 h (Figure 5). Western blots using antibodies against a late golgi integral membrane protease (Kex2p), cytosolic phosphoglycerate kinase (Pgk1p), dolichol phosphate mannose synthase of the ER membrane (Dpm1p) and a component of the nuclear pore complex (Nsp1p) showed equivalent protein contents in all strains. On the other hand, Western blots using antibodies against mitochondrial outer membrane protein Por1p or mitochondrial Atp1p (α_-F1 subunit) revealed that the protein levels were considerably decreased in Δ_fmc1 cells, reaching, respectively, 70 and 52% of the level of proteins in WT. The reason for the discrepancy in the protein degradation is not known. However, in Δ_fmc1_ cells grown under such conditions, F1 sector was not correctly assembled into a functional Fo-F1 ATPase, since ΔΨ collapse prevents Atp1p import into mitochondria; non imported Atp1p might still be trapped into aggregates.20, 21, 22 Such an enrichment of proteins in clusters might provide a clue for a more efficient degradation of Atp1p compared to Por1p. It is nonetheless noteworthy that in both cases, this degradation was largely prevented in the macroautophagy-deficient double mutant Δ_fmc1_ Δ_atg5_.
Figure 5
Selective degradation of mitochondria during autophagy induced by mitochondrial defects. Cells were grown anaerobically for 24 h at 37°C, and 20 μ_g total protein extracts were analysed on a 10% SDS-PAGE. Immunodetection of mitochondria was performed using anti-Atp1p and anti-Por1p antibodies; immunodetection of late golgi, cytosolic, ER and nuclear compartments was performed, respectively, with anti-Kex2p, anti-Pgk1p, anti-Dpm1p, and anti-Nsp1 antibodies. Wild-type (WT), strain MCP1-8; Δ_fmc1 cells, strain MCP6-8; Δ_atg_5 cells, strain MCP185; Δ_fmc1_Δ_atg5_ cells, strain MCP685. Western blot is representative of at least three independent experiments
Taken together, these data indicate that the mitochondrial defect generated by anaerobic growth of mutant cells carrying a nonfunctional F0-F1 ATPase triggers mitophagy through a process that most likely relies on the classical ATG machinery.
Mitochondrial damage-induced autophagy is independent of the ATP synthase structural defect and can be triggered by other mitochondrial mutations or pharmacological treatments mimicking the mitochondrial defects in FMC1 null mutant
In mammalian cells, cytosolic protein aggregates have already been suggested to induce autophagy.29, 30 Furthermore, in yeast, impairment of the Yme1p-mediated degradation of Cox2p subunits that are not correctly assembled in cytochrome c oxidase complex of the respiratory chain, leads to mitochondrial degradation by a poorly characterised process.31, 32 As already stated, Δ_fmc1_ cells grown at nonpermissive temperature are known to accumulate _α_-F1 and β_-F1 aggregates in the mitochondrial matrix.22 A tempting hypothesis would, therefore, be that the autophagic process depicted here could result from a similar signalling initiated by these matricial aggregates. However, Δ_fmc1 cells grown under aerobic conditions at 37°C (Figure 2d) show typical mitochondria harbouring such matricial aggregates (dashed arrows), and yet, autophagy was never detected either by EM (Figure 2d) or with the ALP test (Figure 3, white bar). Hence, protein aggregation in the mitochondrial matrix was not sufficient to initiate mitochondrial damage-induced autophagy.
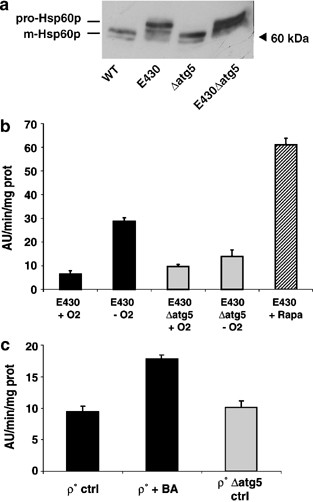
On the other hand, the autophagic activity observed in Δ_fmc1_ cells could result from structural defects in the ATP synthase assembly rather than from the ensuing bioenergetic dysfunction. In order to discriminate between these two hypotheses, the same study was repeated with another F1-deficient mutant (E430), which contains an amino-acid replacement (E222K) in _β_-F1. This point mutation disrupts the catalytic site of ATP synthase and leads to a compromised ATPase activity. However, this mutation does not hinder the assembly of the _α_-F1 and β_-F1 in α_3_β_3 hexamers,33 and therefore, does not induce aggregation of the mutant protein. As in Δ_fmc1 mutant, anaerobic growth of E430 mutant resulted in a defective import of the mitochondrial Hsp60p, which occurred independently of ATG5 disruption, and accounts for ΔΨ collapse (Figure 6a). Under these conditions, E430 cells also showed an increased autophagic activity after 24 h −O2 (Figure 6b, black bars) compared to control conditions (+O2) or to Δ_atg5 mutant in either condition (Figure 6b, grey bars). On the whole, the ALP activity in E430 cells grown anaerobically for 24 or 36 h (not shown) reached 50% of Rapa-stimulated activity (Figure 6b, hatched bar).
Figure 6
Mitochondrial damage-induced autophagy can be stimulated by other mitochondrial mutations and pharmacological treatments mimicking the mitochondrial defects in FMC1 null mutant (a) E430 mutant exhibited alterations in the processing of nuclear-encoded mitochondrial HSP60p when grown anaerobically for 24 h at 28°C. In all, 30 μ_g total protein extracts were analysed on a 9–15% gradient SDS-PAGE. Immunodetection was performed with anti-HSP60 antibody. Wild-type (WT), strain MCP1-8; E430 cells, strain E430 pho8_Δ_60; Δ_atg5 cells, strain MCP185; E430 Δ_atg5_ cells, strain E430 pho8_Δ_60 Δ_atg5_. (b) ALP activity was stimulated in anaerobically grown E430 mutant. ALP activities were measured on the supernatants of permeabilised cells expressing Pho8Δ60p. Black bars: E430 cells (strain E430 pho8_Δ_60); grey bars: E430Δ_atg5_ cells (strain E430 pho8_Δ_60 Δ_atg5_). Cells were grown under aerobiosis (+O2) or anaerobiosis (−O2) for 24 h with or without rapamycin (Rapa, hatched bars). Values presented with the SEM are the average of three independent experiments.(c) ALP activity is stimulated in cells with a pharmacologically-induced mitochondrial defect. Black bars: Rho° cells (strain MCP1-8 Rho°); grey bar Rho°Δ_atg5_ cells (strain MCP185 Rho°). Cells were grown under aerobiosis with or without BA for 14 h. Values presented with the SEM are the average of three independent experiments
Hence, mitochondrial damage-induced autophagy does not result from structural defects in the ATP synthase assembly, but correlates with a functional impairment of the complex and its ensuing bioenergetic dysfunction.
Mitochondrial functional defects (i.e. collapse of ΔΨ) in Δ_fmc1_ and E430 paradigms result, as already stated, from the combination of two parameters: (i) the absence of respiration (due to anaerobiosis) and (ii) the inhibition of ATP hydrolysis (due to an impaired ATPase). These parameters were reproduced in cells grown under normoxi at 28°C using a Rho° strain (nonfunctional respiratory chain) treated with bongkrekic acid (BA), which inhibits the mitochondrial ATP/ADP carrier and thus prevents the entry and further hydrolysis of ATP in the mitochondrial matrix. The ALP activity was assayed in Rho° cells treated for 14 h with BA. Figure 6c shows an increased ALP activity in Rho° cells grown in the presence of BA as compared to untreated cells or to the Δ_atg5_ strain, although the ALP activity reached only 30% of Rapa-stimulated activity (not shown). Similar results were obtained in BA-treated Rho+ cells when respiration was inhibited by the presence of antimycin A, which blocks the complex III of the respiratory chain (not shown). Although these different treatments reproduced the stimuli that initiated autophagy in Δ_fmc1_ and E430 mutants, they appeared to be unsuitable for further studies. Indeed, EM pictures taken at later time points revealed, as expected, the presence of cytosolic autophagosomes in ATG5_-positive cells and their disappearance in Δ_atg5 mutant, but also an unexpected massive vacuolar fragmentation, probably reflecting a side effect of BA on vacuolar homeostasis (not shown).
Mitochondrial damage-induced autophagy does not result from cellular ATP depletion
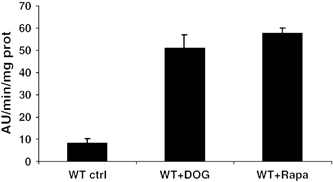
Mitochondrial damage-induced autophagy depicted here was shown to rely on classical ATG machinery (Figure 5); therefore TOR, which is a central regulator of this machinery, could participate in the signal transduction of this process. Recently, TOR-mediated autophagy was reported to be induced in mammalian cells by ATP depletion when glycolysis was inhibited by 2-deoxyglucose (DOG).34 DOG is transported into eukaryotic cells as efficiently as glucose and is converted into 2-DOG 6-phosphate but this phosphorylated form cannot be further metabolised by hexose phosphate isomerase, thus inhibiting glycolysis in early steps. This treatment was reproduced in yeast and DOG was found to stimulate ALP activity as efficiently as Rapa (Figure 7). We thus conclude that glycolysis inhibition, and the ensuing ATP collapse, induced autophagy in yeast also. ATP depletion resulting from DOG addition occurs, in yeast as in mammals, within minutes, whereas the onset of autophagy required at least 1 h; thus ATP collapse is an early event in the signal transduction of a process that appears to be conserved from yeast to mammals.
Figure 7
Effects of DOG on ALP activity. Cells were grown under aerobic conditions in YPD and supplemented with 1% DOG or rapamycin for 13 h. Supernatants of permeabilised cells were assayed for autophagic activity. WT cells (strain MCP1-8). Values presented with the SEM are the average of three independent experiments
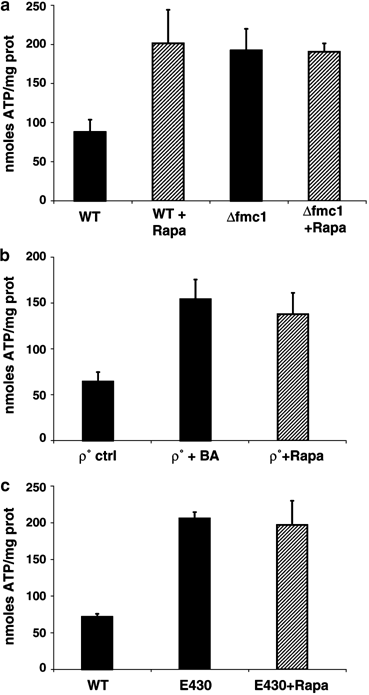
In our experimental models, ATP is no longer hydrolysed by mitochondrial ATP synthase and the cellular ATP balance is consequently also affected. Therefore, we next investigated whether ATP played a role in the signal transduction of the autophagic process described here. Total cellular ATP was thus measured in our three models. Δ_fmc1_ cells grown under autophagy-inducing conditions (37°C, −O2) exhibited a two-fold increase in ATP content as compared to WT cells (Figure 8a, black bars). Similar results were found in Rapa-treated positive controls (Figure 8a, hatched bars). Rho° cells treated with BA also exhibited twice as much ATP as untreated cells (Figure 8b, black bars), as did Rapa-treated cells (Figure 8b, hatched bars). The ATP content in E430 cells cultured under anaerobic conditions for 24 h was increased even further as compared to WT cells (Figure 8c, black bars) and here also attained the level observed in Rapa positive control (Figure 8c, hatched bars). These results suggest that the increase in ATP was associated with autophagy.
Figure 8
Cellular ATP assays. ATP content was measured on supernatants of total cellular extracts from rapamycin-treated (hatched bars) or untreated cells (black bars). The values presented with the SEM are the average of three independent experiments. (a) Cells were pregrown under aerobic conditions in YPD for 12 h at 37°C and shifted to anaerobiosis for 24 h. WT, strain MCP1-8; Δ_fmc1_ cells, strain MCP6-8. (b) Rho° cells (strain MCP1-8 Rho°) were grown under aerobiosis for 14 h. (c) Cells were grown under anaerobiosis for 24 h. WT, strain MCP1-8; E430 cells, strain E430 pho8_Δ_60
Hence, the cellular ATP content and the ALP activities vary accordingly. We thus conclude that the contribution of cellular ATP collapse in the induction of mitochondrial damage-induced autophagy can be ruled out.
Discussion
In this study, we have analysed in yeast the effects of mutations and pharmacological treatments, which impair the bioenergetic status and the biogenesis of mitochondria. These situations correlate with the onset of autophagy, as monitored by EM analysis and enzymatic assays. Autophagy is widely accepted as one of the two major degradative pathways in eukaryotic cells together with the proteasome, and being in charge of long- and short-lived proteins, respectively. Since autophagy has so far mainly been described to be induced by nutrient limitation, it was essentially considered to be an adaptive response in charge of supplying cells with macromolecules during adverse environmental conditions. The results presented here clearly indicate that autophagy surpasses such a function since the process is triggered in our system under nonstarving conditions, and as a response to an intracellular signalling of bioenergetic dysfunction. Our data, therefore, support the emerging concept (reviewed in Ogier-Denis and Codogno35) of an alternative role for autophagy, which envisages the process as a ‘stress-induced’ housekeeping mechanism involved in the general maintenance of cellular homeostasis. Autophagic response was already reported to be induced by misfolded proteins.29, 30 However, if the Δ_fmc1_ model used here effectively accumulates _α_-F1 and _β_-F1 as large matricial aggregates under autophagy-promoting conditions20, 21 (heat stress and anaerobiosis), such aggregates also accumulate in cells grown under aerobiosis (Figure 2d), without induction of autophagy. Hence, protein misfolding is not the ‘stress-inducing’ event of autophagy in our study.
In all the models tested in this work, cells were grown under conditions where they essentially meet their ATP requirement through glycolysis. Although mitochondria do not significantly contribute to ATP production in these cases, all the data concur that mitochondria host vital processes other than respiration and energy supply, the disruption of which triggers autophagy and culminates in cell death (Figure 4). These processes (e.g. synthesis of lipids, heme, amino acids, and nucleotides) depend on the maintenance of ΔΨ for proper mitochondrial protein import,23, 36 which is compromised in our paradigm (Figure 1 and 5a). We propose that impairment of mitochondrial biogenesis is the source of the autophagic process described here.
Although autophagy is generally acknowledged as a nonspecific degradative process capable of sequestering bulk cytoplasm as well as entire organelles into autophagosomes, the concept of selective autophagy has recently been documented by a growing number of reports. As a consequence, the housekeeping function assigned to autophagy has broadened to the maintenance of organelle turnover. In yeast, selective microautophagy targets nonessential components of the nucleus during PMN;13 peroxisomes are also eliminated by pexophagy upon modification of the carbon source;11 finally, autophagy also mediates the early removal of fully functional mitochondria when yeast cells are shifted from a respiratory metabolism (i.e. a respiratory substrate will be metabolised by mitochondria to provide the energetic supply) to a fermentative metabolism (i.e. energy is supplied by sequential degradation of a fermentative substrate through glycolysis).37 In this latter case, mitophagy clearly reflects another physiological adaptive role played by autophagy, which is, however, still nutrition-related. We found that preferential degradation of mitochondria could be prompted in response to mitochondrial damage and therefore, we conclude that stress-induced mitophagy is another asset of autophagy physiology in yeast. Whether this process is reversible – as classical autophagy – or not remains unclear. We found that mitophagy correlates with cell death and further work will determine if cells can recover from situations compromising mitochondria, or if autophagy could be envisaged as an ultimate programme initiated when cells can no longer recover from mitochondrial injuries. Such a finding would raise the attractive proposal that autophagy could be turned from a survival programme into a cell death mechanism in yeast as well.
As a matter of fact, a similar description of mitophagy preceding cell death was observed in mammals, where selective removal of mitochondria was reported to target nonfunctional organelles when the apoptotic programme was blocked at a postmitochondrial stage14, 17, 18, 19 (after the release of apoptogenic factors from the organelles), or when mitochondria carry deleterious DNA mutations.38 Whether this mitochondrial degradation results from direct lysosomal uptake18, 38 or from autophagosomal sequestration15, 17 is still unclear, but under these conditions, mitophagy would control the scavenging of damaged mitochondria. However, the role devoted to mitophagy in the process ultimately leading to cell death is still unknown.
Molecular characterisation of mitophagy still awaits the identification of the machinery involved (i) in the signal transduction of the process, and (ii) in the selectivity of mitochondria. Such information will be required to determine whether or not nutrient-induced mitophagy39 relies on the same molecular mechanisms as mitochondrial damage-induced mitophagy. Pioneering work in yeast recently identified Uth1p as the first mitochondrial protein required for early removal of fully functional mitochondria upon modification of the carbon source.37, 39 Whether Uth1p is required for mitochondrial damage-induced mitophagy is currently being evaluated. As in nutrient-induced mitophagy,37, 39 we found that mitochondrial damage-induced mitophagy was largely prevented when macroautophagy was disrupted (Figure 5); thus, mitophagy most likely involves TOR signalling; but knowing whether microautophagy participates in the process, as suggested by EM data (Figure 1), will require further work. Our attempt to gain insights into the signal transduction of mitophagy resulted in the intriguing finding that the cellular ATP content increased during the process. Such a phenomenon has never been documented before, at least not to our knowledge, and the underlying molecular mechanism is still unknown. Nevertheless, the fact that ATP variation is comparable in our models and in Rapa-treated cells is a good indication that both are relevant in the unique cellular process that is autophagy. This observation is far from being counter-intuitive considering the fact that most ATP-consuming processes (transcription/translation) are blocked during autophagy, and that the general break down of dispensable macromolecules aids cellular survival. Further studies are needed to decipher the role and fate of ATP in this process.
Autophagy has, to date, been mostly known as a starvation-induced cellular response designed to face nutrient limitation, and eventually leading to cell death if taken to completion. With respect to the data presented here, the physiological relevance of mitochondrial damage-induced autophagy gains credit and the fact that the process is conserved from yeast to mammals will undoubtedly help in characterising the underlying molecular mechanisms. In the light of our work, autophagy can be envisaged as a quality-control process aimed to prevent proliferation of defective mitochondria, and may even turn out to be a death programme initiated when cells cannot recover from mitochondrial damage. Our work, therefore, provides grounding for the emerging view that failures of the autophagic programme in cells could account for the appearance of mitochondrial pathologies, in the early stages of life as well as in aged cells. Indeed, most mitochondrial pathologies (neurodegenerative disorders, myopathies, diabetes, etc.) are related to defects in oxidative phosphorylations, and our results point toward autophagy as a new target to prevent the emergence of such pathologies.
Materials and Methods
Strains and media
Yeast strains in this study were constructed with the use of standard yeast genetic methods, and are listed in Table 1.
Table 1 Strains used in this study
Deletion of the ATG5 gene was generated in MC1, MC6, and E430 strains by replacement of the gene with a kanamycin cassette (LoxP-kanMX-LoxP) PCR-amplified from plasmid pUG640 using the following primers: Atg5-Kan, 5′-CAT TAA ACA ATT ACT TTG GAA TGG TGA GCT TAA TGT GCA TAG GCC ACT AGT GGA TCT G-3′; Kan-Atg5, 5′-TTA GAG CTC AGA GGA AGC TTT ATC GCC GCC TTT TAT AGG AAC AAG CAG CTG AAG CTT CGT ACG C-3′. The PHO8 gene in each of these strains was replaced with pho8_Δ_60 by insertion of the plasmid pTN9 (a gift from Y Ohsumi) into PHO8 locus and excision out as described previously.28 Rho° cells were generated by a two-step growth of the parental strains on YPD plates supplemented with ethidium bromide (40 _μ_g/ml). Growth of yeast cells under anaerobiosis was as previously described.25 Briefly, cells were grown in liquid YPDA medium containing 30 _μ_g ml−1 ergosterol (Sigma) and 0.2% Tween 80 (Sigma). Anaerobiosis was achieved by sealing six-well plates and Anaerocult P system (Merck) in special incubation bag.
BA (10 _μ_M) and 2-DOG (1%) were from Sigma. Rapamycin (0,38 _μ_M) was from Alexis.
Western blot
Total protein extracts were separated on SDS-PAGE, transferred onto a PVDF membrane (Millipore) and revealed with ECL (Amersham). Primary antibodies were used at 1/2000 dilution. Anti-Hsp60p antibody was from Imgenex Corporation. Anti-Pgk1p, anti-Por1p, anti-Dpm1p, and anti-Nsp1 antibodies were gifts from Dr S Manon (IBGC, Bordeaux, France), anti-Atp1p was a gift from Dr J Velours (IBGC, Bordeaux, France) and anti-Kex2p was a gift from Dr S Nothwehr (University of Missouri, MO, USA).
ALP assay
Cells were harvested at OD550=2. The assay method performed on permeabilised whole cells using _α_-naphthyl phosphate (Sigma) as substrate was as previously described.41 Fluorescence intensity was measured at 472 nm after excitation at 345 nm. Protein concentration was determined using BCA (Pierce).
Freezing and freeze-substitution for ultrastructural studies
The yeast pellets were placed on the surface of formvar-coated copper EM grids (400 mesh). Each loop was quickly submersed in liquid propane (−180°C) and then transferred to a precooled solution of 4% osmium tetroxide in dry acetone at −82°C for 48 h for substitution fixation. Samples were gradually warmed up to room temperature, and washed in dry acetone. Specimens were stained for 1 h in 1% uranyl acetate in acetone 4°C, rinsed and infiltrated with araldite (epoxy resin, Fluka). Ultra-thin sections were stained with lead citrate and were viewed on a Philips Tecnai 12 Biotwin (120 kV) electron microscope (SERCOMI, Université Victor Segalen Bordeaux 2).
Cellular ATP assay
Cells were harvested at OD550=3 and extraction of total proteins was performed with 5% TCA. Protein assays were performed on acetone-washed pellets with BCA (Pierce). Supernatants were neutralised with KOMO (MOPS 0.3 M, KOH 2 M) and luminescence was measured on an aliquot with Quantitative ATP Monitoring Kit from Thermo Labsystems on a Victor luminometer (Perkin Elmer).
Abbreviations
ALP:
alkaline phosphatase
BA:
bongkrekic acid
ΔΨ:
mitochondrial electrochemical transmembrane potential
DOG:
2-deoxyglucose
EM:
electron micrographs
PCD:
programmed cell death
Rapa:
rapamycin
WT:
wild type
References
- Takeshige K, Baba M, Tsuboi S, Noda T and Ohsumi Y (1992) Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119: 301–311
Article CAS Google Scholar - Schworer CM and Mortimore GE (1979) Glucagon-induced autophagy and proteolysis in rat liver: mediation by selective deprivation of intracellular amino acids. Proc. Natl. Acad. Sci. USA 76: 3169–3173
Article CAS Google Scholar - Bursch W, Ellinger A, Gerner C, Frohwein U and Schulte-Hermann R (2000) Programmed cell death (PCD). Apoptosis, autophagic PCD, or others? Ann. NY Acad. Sci. 926: 1–12
Article CAS Google Scholar - Crespo JL and Hall MN (2002) Elucidating TOR signaling and rapamycin action: lessons from Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 66: 579–591
Article CAS Google Scholar - Klionsky DJ, Cregg JM, Dunn WA, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M and Ohsumi Y (2003) A unified nomenclature for yeast autophagy-relayed genes. Dev. Cell 5: 539–545
Article CAS Google Scholar - Darsow T, Rieder SE and Emr SD (1997) A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J. Cell Biol. 138: 517–529
Article CAS Google Scholar - Sato TK, Darsow T and Emr SD (1998) Vam7p, a SNAP-25-like molecule, and Vam3p, a syntaxin homolog, function together in yeast vacuolar protein trafficking. Mol. Biol. Cell 18: 5308–5319
Article CAS Google Scholar - Baba M, Takeshige K, Baba N and Ohsumi Y (1994) Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J. Cell Biol. 124: 903–913
Article CAS Google Scholar - Dean RT (1977) lysosomes and membrane recycling. A hypothesis. Biochem. J. 168: 603–605
Article CAS Google Scholar - Sakai Y, Koller A, Rangell LK, Keller GA and Subramani S (1998) Peroxisome degradation by microautophagy in Pichia pastoris : identification of specific steps and morphological interlediates. J. Cell Biol. 141: 625–636
Article CAS Google Scholar - Tuttle DL and Dunn WA (1995) Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris. J. Cell Sci. 108: 25–35
CAS PubMed Google Scholar - Muller O, Sattler T, Flotenmeyer M, Schwarz H, Plattner H and Mayer A (2000) Autophagic tubes: vacuolar invaginations involved in lateral membrane sorting and inverse vesicle budding. J. Cell Biol. 151: 519–528
Article CAS Google Scholar - Roberts P, Moshitch-Moshkovitz S, Kvam E, O'Toole E, Winey M and Goldfarb DS (2003) Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell 14: 129–141
Article CAS Google Scholar - Lemasters JJ, Nieminen AL, Qian T, Trost LC, Elmore SP, Nishimura Y, Crowe RA, Cascio WE, Bradham CA, Brenner DA and Herman B (1998) The mitochondrial permeability transition in cell death: a common mechanism in necrosis, apoptosis and autophagy. Biochim. Biophys. Acta. 1366: 177–196
Article CAS Google Scholar - Tolkovsky AM, Xue L, Fletcher GC and Borutaite V (2002) Mitochondrial disappearance from cells: a clue to the role of autophagy in programmed cell death and disease? Biochimie. 84: 233–240
Article CAS Google Scholar - Xue L, Fletcher GC and Tolkovsky AM (1999) Autophagy is activated by apoptotic signalling in sympathetic neurons: an alternative mechanism of death execution. Mol. Cell Neurosci. 14: 180–198
Article CAS Google Scholar - Xue L, Fletcher GC and Tolkovsky AM (2001) Mitochondria are selectively eliminated from eukaryotic cells after blockade of caspases during apoptosis. Curr. Biol. 11: 361–365
Article CAS Google Scholar - Elmore SP, Qian T, Grissom SF and Lemasters JJ (2001) The mitochondrial permeability transition initiates autophagy in rat hepatocytes. FASEB J. 15: 2286–2287
Article CAS Google Scholar - Bernard D, Slomianny C, Vandenbunder B and Abbadie C (2001) cRel induces mitochondrial alterations in correlation with proliferation arrest. Free Radic. Biol. Med. 31: 943–953
Article CAS Google Scholar - Lefebvre-Legendre L, Vaillier J, Benabdelhak H, Velours J, Slonimski PP and di Rago JP (2001) Identification of a nuclear gene (FMC1) required for the assembly/stability of yeast mitochondrial F(1)-ATPase in heat stress conditions. J. Biol. Chem. 276: 6789–6796
Article CAS Google Scholar - Ackerman SH and Tzagoloff A (1990) Identification of two nuclear genes (ATP11, ATP12) required for assembly of the yeast F1-ATPase. Proc. Natl. Acad. Sci. USA 87: 4986–4990
Article CAS Google Scholar - Lefebvre-Legendre L, Salin B, Schaëffer J, Brèthes D, Dautant A, Ackerman SH and di Rago JP (2005) Failure to assemble the _α_3_β_3 subcomplex of the ATP synthase leads to accumulation of the α and β subunits within inclusion bodies and the loss of mitochondrial cristae in Saccharomyces cerevisiae. J. Biol. Chem. 280: 18386–18392
Article CAS Google Scholar - Gbelska Y, Subik J, Svoboda A, Goffeau A and Kovac L (1983) Intramitochondrial ATP and cell functions: yeast cells depleted of intramitochondrial ATP lose the ability to grow and multiply. Eur. J. Biochem. 130: 281–286
Article CAS Google Scholar - Neupert W (1997) Protein import into mitochondria. Ann. Rev. Biochem. 66: 863–917
Article CAS Google Scholar - Lefebvre-Legendre L, Balguerie A, Duvezin-Caubet S, Giraud MF, Slonimski PP and Di Rago JP (2003) F1-catalysed ATP hydrolysis is required for mitochondrial biogenesis in Saccharomyces cerevisiae growing under conditions where it cannot respire. Mol. Microbiol. 47: 1329–1339
Article CAS Google Scholar - Kametaka S, Matsuura A, Wada Y and Ohsumi Y (1996) Structural and functional analyses of APG5, a gene involved in autophagy in yeast. Gene 178: 139–143
Article CAS Google Scholar - George MD, Baba M, Scott SV, Mizushima N, Garrison BS, Ohsumi Y and Klionsky DJ (2000) Apg5p functions in the sequestration step in the cytoplasm-to-vacuole targeting and macroautophagy pathways. Mol. Biol. Cell 11: 969–982
Article CAS Google Scholar - Noda T, Matsuura A, Wada Y and Ohsumi Y (1995) Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 210: 126–132
Article CAS Google Scholar - Ravikumar B, Duden R and Rubinsztein DC (2002) Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 11: 1107–1117
Article CAS Google Scholar - Fortun J, Dunn WA, Joy S, Li J and Notterpek L (2003) Emerging role for autophagy in the removal of aggresomes in Schwann cells. J. Neurosci. 23: 10672–10680
Article CAS Google Scholar - Campbell CL and Thorsness PE (1998) Escape of mitochondrial DNA to the nucleus in yme1 yeast is mediated by vacuolar-dependent turnover of abnormal mitochondrial compartments. J. Cell Sci. 111 (Part 16): 2455–2464
CAS PubMed Google Scholar - Shafer KS, Hanekamp T, White KH and Thorsness PE (1999) Mechanisms of mitochondrial DNA escape to the nucleus in the yeast Saccharomyces cerevisiae. Curr. Genet. 36: 183–194
Article CAS Google Scholar - Liang L and Ackerman SH (1996) Characterization of mutations in the β subunit of the mitochondrial F1-ATPase that produce defects in enzyme catalysis and assembly. J. Biol. Chem. 271: 26522–26528
Article CAS Google Scholar - Dennis PB, Jaeschke A, Saitoh M, Fowler B, Kozma SC and Thomas G (2001) Mammalian TOR: a homeostatic ATP sensor. Science 294: 1102–1105
Article CAS Google Scholar - Ogier-Denis E and Codogno P (2003) Autophagy: a barrier or an adaptive response to cancer. Biochim. Biophys. Acta 1603: 113–128
CAS PubMed Google Scholar - Visser W, Van der Baan A, Batenburg-van der Vegte W, Scheffers WA, Kräemer R and Van Dijken JP (1994) Involvement of mitochondria in the assimilatory metabolism of anaerobic Saccharomyces cerevisiae cultures. Microbiology 140: 3039–3046
Article CAS Google Scholar - Kissova I, Deffieu M, Manon S and Camougrand N (2004) Uth1p is involved in the autophagic degradation of mitochondria. J. Biol. Chem. 279: 39068–39074
Article CAS Google Scholar - Gu Y, Wang C and Cohen A (2004) Effect of IGF-1 on the balance between autophagy of dysfunctional mitochondria and apoptosis. FEBS Lett. 577: 357–360
Article CAS Google Scholar - Camougrand N, Kissova I, Velours G and Manon S (2004) Uth1p: a yeast mitochondrial protein at the crossroads of stress, degradation and cell death. FEMS Yeast Res. 5: 133–140
Article CAS Google Scholar - Güldener U, Heck S, Fielder T, Beinhauer J and Hegemann J (1996) A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24: 2519–2524
Article Google Scholar - Nothwehr SF, Bryant NJ and Stevens TH (1996) The newly identified yeast GRD genes are required for retention of late-Golgi membrane proteins. Mol. Cell Biol. 16: 2700–2707
Article CAS Google Scholar
Acknowledgements
We thank Dr Oliver for critical reading of the manuscript and Dr Manon for many helpful discussions. We also thank Dr Ohsumi, Dr Noda, Dr Manon, and Dr Ackerman for providing biological material. This work was supported by a Roche Research Foundation fellowship (to MP), funded by Swiss National Science Foundation grant 3100-061380 and AFM grant (to JPdR).
Author information
Authors and Affiliations
- Département de Biologie Cellulaire, Université de Genève, Suisse
M Priault & J-C Martinou - IBGC/CNRS, Bordeaux cedex, France
B Salin, J Schaeffer & J-P di Rago - INSERM UMR 601, Nantes cedex, France
F M Vallette
Authors
- M Priault
You can also search for this author inPubMed Google Scholar - B Salin
You can also search for this author inPubMed Google Scholar - J Schaeffer
You can also search for this author inPubMed Google Scholar - F M Vallette
You can also search for this author inPubMed Google Scholar - J-P di Rago
You can also search for this author inPubMed Google Scholar - J-C Martinou
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toM Priault.
Additional information
Edited by E Baehrecke Margherita
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Supplementary information
Rights and permissions
About this article
Cite this article
Priault, M., Salin, B., Schaeffer, J. et al. Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast.Cell Death Differ 12, 1613–1621 (2005). https://doi.org/10.1038/sj.cdd.4401697
- Received: 15 December 2004
- Revised: 05 April 2005
- Accepted: 27 May 2005
- Published: 10 June 2005
- Issue Date: 01 December 2005
- DOI: https://doi.org/10.1038/sj.cdd.4401697