Combined Application of NMR- and GC-MS-Based Metabonomics Yields a Superior Urinary Biomarker Panel for Bipolar Disorder (original) (raw)
Introduction
Bipolar disorder (BD) is one of the top ten most disabling disorders in working age adults and affects up to 1% of the general population1,2. Due to the lack of objective diagnostic modalities, the diagnosis of BD still relies on the subjective identification of symptomatic clusters3. However, the clinical symptoms of BD are considerably complex and diverse3, resulting in a high rate of underdiagnosis and misdiagnosis that contributes to increased suicide risk and poorer prognosis4. Given these facts, there is an urgent need to identify objective laboratory-based diagnostic biomarkers for BD.
Metabonomics – the comprehensive analysis of low-molecular-weight endogenous metabolites in a biological sample – has been widely applied to capture the metabolic changes in various disease states5. Currently, there are three major analytical techniques that are suited for non-targeted metabonomic mapping: nuclear magnetic resonance (NMR) spectroscopy, gas chromatography-mass spectroscopy (GC-MS) and liquid chromatography- mass spectroscopy (LC-MS)6,7,8. Each of these analytical techniques has its advocates and possess their own unique features. An increasing number of researchers have applied these techniques to identify diagnostic biomarkers for neuropsychiatric disorders, including stroke, multiple sclerosis, schizophrenia and autism9,10,11,12,13. Using NMR and GC-MS, our group has successfully identified several potential metabolite biomarkers in the plasma and urine of major depressive disorder (MDD) patients, which could effectively distinguish depressed subjects from healthy controls14,15,16.
With regards to BD, previous metabonomic studies have used a NMR metabonomic platform to identify differential metabolites in post-mortem brain tissue and plasma17,18. Meanwhile, in our group, Zheng et al. employed NMR to identify four potential urinary metabolite biomarkers and Xu et al. employed GC-MS to identify 2,4-dihydroxypyrimidine as a potential urinary metabolite biomarker for diagnosing BD19,20. These previous metabonomic studies have been helpful in developing objective laboratory-based testing for BD while providing valuable data on the physiopathologic mechanism(s) of BD. However, one limitation shared by all these studies was that the researchers only used one metabonomic platform. Irrespective of the unique advantages of any particular methodology, no single metabonomic platform can provide adequate coverage of the entire human metabonome in any given biological sample21. Previous studies have demonstrated that the use of multiple metabolomics platforms and technologies allowed us to identify several previously unknown urine metabolites and to substantially enhance the level of metabolome coverage22,23,24. Therefore, the combined application of NMR spectroscopy and GC-MS may identify a more comprehensive metabolite panel than any single metabonomic platform alone.
Here, in order to investigate the complementary nature of NMR spectroscopy and GC-MS for metabonomic analysis, a novel urinary metabolite panel for diagnosing BD was constructed using a dual platform approach (NMR spectroscopy and GC-MS). The diagnostic performance of the current composite biomarker panel was then comparatively assessed against the previous single platform-derived metabolite panels.
Results
Univariate analysis
Prior to analysis, data was scaled to unit variance. We did univariate analysis using all subjects to find metabolites that would be worthy of further analysis (p < 0.10), which identified 67 different metabolites (18 NMR-derived and 49 GC-MS-derived metabolites) from 94 metabolites. These 94 differential metabolites including the four metabolites biomarkers (choline, N-methylnicotinamide,α-hydroxybutyrate, isobutyrate) identified by NMR and one metabolite (2,4-dihydroxypyrimidine) identified by GC/MS were included in this study19,20. The 94 metabolites were described in supplementary Table S1. And a typical NMR and GC-MS spectrum was described in supplementary figure S1.
OPLS-DA model
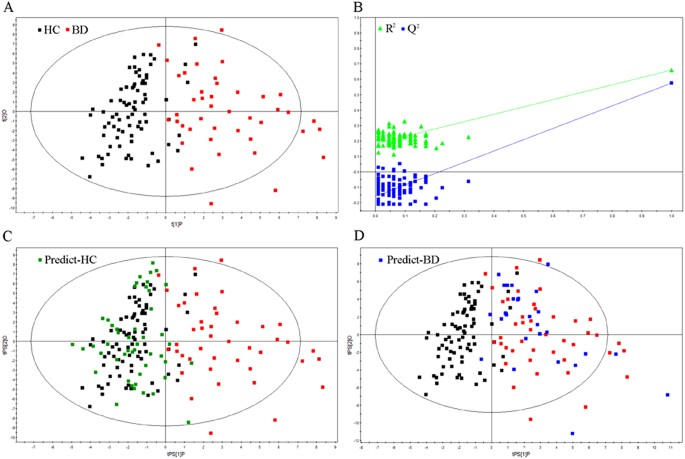
OPLS-DA analysis was carried out to explore the metabolic differences between BD subjects and healthy controls. The 67 differential metabolites were used to perform OPLS-DA analysis. In the training set, the score plots of the OPLS-DA model showed that the BD subjects were obviously separated from healthy controls with little overlap (R2X cum = 0.36, R2Y cum = 0.66, Q2 = 0.57; Figure 1a). The values of those parameters quantifying OPLS-DA model were positive, demonstrating a robust metabolic difference between BD subjects and healthy controls. Furthermore, the permutation test showed the constructed OPLS-DA model was positive and valid (Figure 1b).
Figure 1
Metabonomic analysis of urine samples.
(a) Orthogonal partial least-squares discriminant analysis (OPLS-DA) score plots showing a clear discrimination between BD subjects (red box) and healthy controls (black box) in the training set. (b) Permutation test showing the original R2 and Q2 values (top right) as significantly higher than the corresponding permuted values (bottom left), demonstrating the OPLS-DA model's robustness. (c) OPLS-DA model was used to predict the healthy controls (green box) from the test set. (d) OPLS-DA model was used to predict the BD subjects (blue box) from the test set.
In the test set, 28 BD subjects and 48 healthy controls were used to independently validate the diagnostic performance of the OPLS-DA model. Consequently, samples were correctly predicted by the OPLS-DA model, yielding a predictive accuracy of 96.1% (Figure 1c, d).
Differential metabolites
The coefficient loading plots of the OPLS-DA model identified 33 differential metabolites (|r| > 0.301) responsible for distinguishing BD subjects from healthy controls. Meanwhile, the corresponding OPLS-DA loading plots identified 27 differential metabolites (VIP > 1.0) responsible for discriminating the two groups. Finally, we identified 26 differential metabolites with a VIP > 1.0 and |r| > 0.301 (Table 1). These metabolites included three previously identified biomarkers (α-hydroxybutyrate, N-methylnicotinamide and 2,4-dihydroxypyrimidine)19,20.
Table 1 Key metabolites responsible for discriminating BD subjects from HC
Logistical regression model
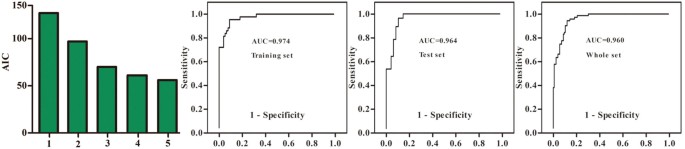
In order to identify a simplified metabolite biomarker panel for BD diagnosis, the 26 differential metabolites were used to perform forward stepwise regression analysis. The AIC rule was applied for model selection. The results demonstrated that the most significant deviations between BD subjects and healthy controls could be described by the combination of four GC-MS-derived metabolites (β-alanine, 2,4-dihydroxypyrimidine, azelaic acid and pseudouridine) and one NMR-derived metabolite (α-hydroxybutyrate) (Figure 2). β-alanine, azelaic acid and α-hydroxybutyrate were increased, while pseudouridine and 2,4-dihydroxypyrimidine, were decreased in BD subjects. The model yielded an average accuracy of 90.1% in the training set and a predictive accuracy of 82.3% in the test set. In the whole set, the model yielded an average accuracy of 88.3%.
Figure 2
Identification and validation of urinary metabolite panel.
Akaike information criterion (AIC) of each model was presented. The current model constructed with five urinary metabolites (2,4-dihydroxypyrimidine, azelaic acid, β-alanine, pseudouridine and α-hydroxybutyrate) showed the highest predictive ability. Receiver operating characteristic (ROC) curve analysis was performed to evaluate the diagnostic performance of this five-biomarker panel. Area under the curve (AUC) values of the training set, test set and whole set were 0.974, 0.964 and 0.960, respectively.
ROC curve analysis
Receiver-operating characteristic (ROC) curve analysis was further performed to quantify the diagnostic performance of this model in the training set, test set and whole set. The area under the curve (AUC) was 0.974 in the training set, 0.964 in the test set and 0.960 in the whole set (Figure 2). The cut-off value of training set, test set and whole set was 0.508, 0.537 and 0.525, respectively. The sensitivity of training set, test set and whole set was 0.860, 0.964 and 0.833, respectively. The specificity of training set, test set and whole set was 0.923, 0.875 and 0.913, respectively. The diagnostic performance of this model constructed with only five differential metabolites is similar to the OPLS-DA model constructed with all 67 differential metabolites, demonstrating the efficacy of this simplified urinary metabolite panel in diagnosing BD.
Combined platform panel versus single platform panel
All BD subjects were used to compare the diagnostic efficacy of the panel found by the combined analytical platforms with the panels found by each single analytical platform. The two cut-off values were from our previous studies19,20. Finally, we obtained the following results: (i) 22 and 19 samples were wrongly predicted by NMR-derived panel and GC-MS-derived panel, respectively; (ii) 12 samples were wrongly predicted by the combined panel; (iii) 10/13 and 9/10 samples were wrongly predicted by the NMR-derived panel and the GC-MS-derived panel, respectively, but were correctly predicted by the combined panel; (iv) 6/9 samples were wrongly predicted both by the NMR-derived panel and the GC-MS-derived panel were also wrongly predicted by the combined panel; and (v) 37/39 samples were correctly predicted by the NMR-derived panel and the GC-MS-derived panel were also correctly predicted by the combined panel.
We also compared the panel derived in this work (β-alanine, 2,4-dihydroxypyrimidine, azelaic acid, pseudouridine and α-hydroxybutyrate) with a composite panel directly generated by the five biomarkers identified in our previous studies19,20. The specificity of the two panels were comparable (current panel 91.3% versus previous panel 90.5%), but the sensitivity of the current panel was higher (current panel 83.1% versus previous panel 71.8%).
Discussion
BD is a common and debilitating mental disorder. However, there is still no empirical laboratory-based test for BD to facilitate its diagnosis. Over the past few years, our research group has focused on addressing this issue. Zheng et al. has found four potential metabolite biomarkers by a NMR-based analytical platform and Xu et al. has found one metabolite biomarker by a GC-MS-based analytical platform. However, due to the diverse physicochemical properties and wide concentration ranges of metabolites21, a single analytical technology cannot provide complete coverage of the human metabonome. Therefore, here we applied a dual platform that generated a panel of five potential metabolite biomarkers (one from NMR, four from GC-MS). The model yielded an average accuracy of 90.1% in the training set and a predictive accuracy of 82.3% in the test set. In the whole set, the model yielded an average accuracy of 88.3%. After comparing the current panel (β-alanine, 2,4-dihydroxypyrimidine, azelaic acid, pseudouridine and α-hydroxybutyrate) with a composite panel generated by the five biomarkers identified in our previous studies (2,4-dihydroxypyrimidine, N-methylnicotinamide, choline, isobutyrate and α-hydroxybutyrate), the specificity of the two panels were comparable (current panel 91.3% versus previous panel 90.5%), but the sensitivity of the current panel was higher (current panel 83.1% versus previous panel 71.8%). Additionally, among the thirteen BD samples wrongly predicted by NMR but correctly predicted by GC-MS, ten samples were correctly predicted by the current panel and among the ten BD samples wrongly predicted by GC-MS but correctly predicted by NMR, nine samples were correctly predicted by the current panel. These results demonstrate the complementary nature of NMR spectroscopy and GC-MS for metabonomic analysis and indicate that the combined application of NMR and GC-MS platforms should provide a better strategy for identifying meaningful biomarkers than relying on any single technique in isolation.
Among the five metabolite biomarkers identified here, α-hydroxybutyrate and 2,4-dihydroxypyrimidine were also identified in our previous studies19,20. α-hydroxybutyrate was increased in BD patients. Zheng et al. reported that higher urinary α-hydroxybutyrate suggests increased oxidative stress in BD patients19 and Andreazza et al. reported that increased oxidative stress is associated with the pathophysiology of BD25. 2,4-dihydroxypyrimidine was decreased in BD patients. Due to its involvement in glutamine formation26, the significantly decreased level of 2,4-dihydroxypyrimidine implies that disturbance of glutamine was involved in the onset of BD20. In agreement with this speculation, many previous metabonomic analyses of post-mortem brain tissue and plasma obtained from BD patients have also observed significant alterations in glutamine levels17,18.
Additionally, β-alanine, azelaic acid and pseudouridine were identified as metabolite biomarker here. Xu et al. reported that only azelaic acid and pseudouridine, but not β-alanine, were differential metabolites between BD subjects and healthy controls20. Meanwhile, Zheng et al. identified choline, isobutyrate and N-methylnicotinamide19, none of which were identified in the current work. Considering the higher accuracy of the current panel, these results show the advantage of a dual platform approach in detecting the sometimes subtle metabolic differences between experimental groups. Therefore, the combined use of these two techniques is recommended for future metabonomic studies to better enable the discovery of novel diagnostic biomarkers.
When we only used the GC/MS metabolites to identify potential metabolite biomarker panel, seven metabolites (24dihydroxypyrimidine, aminoethanol, arabitol, RS23dihydroxybutanoicacid, phenylalanine azelaic acid, β-alanine) were included into the potential diagnostic model. And the discrimination power of this model was comparable to the model consisting of the four GC/MS metabolites (β-alanine, 2,4-dihydroxypyrimidine, azelaic acid and pseudouridine) in the current biomarker panel. Meanwhile, the sensitivity, specificity and AUC of the four GC/MS metabolites were all lower than that of the current biomarker panel. These results suggested that the combined application of NMR- and GC-MS-Based metabonomics could identify the most typical and representative metabolites panel than applying each platform in isolation.
To analyze the biological functions of these identified potential metabolite biomarkers, the pathway analysis was performed by online software MetaboAnalyst27. These metabolites were mainly involved in four metabolic pathways (Supplementary Table S2). About these metabolic pathways, previous study has found that genetically determined aberrations in pyrimidine metabolism were associated clinically with various degrees of mental retardation and/or unexpected and often devastating neurological dysfunction28. Pantothenate and CoA biosynthesis was found to be unique to major depression in a study exploring thalamic transcriptome screening in three psychiatric states29, but, in the current work, the results indicated that it might be affected in patients with BD. Beta-alanine appeared to act via glycine and GABA (A) receptors (both inhibitory neurotransmitters) with comparable efficacy to glycine and GABA themselves30. And, expression of GABA (A) receptors was altered significantly in the lateral cerebellum of subjects with BD31. In a study listing potential targets for novel therapeutics for BD, one of the suggested targets was a glutamate propionic acid receptor32, which is part of the propanoate metabolism pathway.
A few limitations of this study should be addressed here. First, a relatively small sample size of non-medicated BD subjects was recruited. Larger samples are needed to determine the influences of drugs upon these identified biomarkers. Second, these biomarkers were confirmed solely by comparing BD patients with healthy controls. Therefore, it was unknown whether or not there biomarkers can be effectively used to discriminate BD from other psychiatric disorders that can present in a similar fashion, such as depression and schizophrenia. Third, future studies should collect cerebrospinal fluid (CSF) from BD patients to ensure that these urinary biomarkers are physiologically relevant to disease pathogenesis. Finally, although a dual platform approach was used here, these biomarkers still require verification through additional metabonomic methods such as LC-MS.
In conclusion, using a dual platform metabonomic approach, a novel composite urinary metabolite biomarker panel for diagnosing BD was identified. This panel, consisting of one metabolite detected by NMR (α-hydroxybutyrate) and four metabolites detected by GC-MS (2, 4-dihydroxypyrimidine, azelaic acid, β-alanine and pseudouridine), could differentiate BD subjects from healthy controls with a higher accuracy than our previous single platform-derived metabolite biomarker panels. Thus, the urinary biomarker panel identified here shows promise as an effective diagnostic tool for BD. These findings also demonstrate the complementary nature of NMR spectroscopy and GC-MS for metabonomic analysis, suggesting that the combination of NMR spectroscopy and GC-MS can identify a more comprehensive metabolite panel than applying each platform in isolation.
Methods
Subjects & sample collection
Prior to sample collection, written informed consents were obtained from all recruited subjects. The protocols of this study were reviewed and approved by the Ethical Committee of Chongqing Medical University. The methods were carried out in accordance with the approved guidelines and regulations. BD subjects were recruited from the psychiatric center of the First Affiliated Hospital at Chongqing Medical University (Chongqing, China). The candidates had to meet the following inclusion criteria: (i) a diagnosis of BD according to the Structured Clinical Interview from the DSM-IV-TR; (ii) no pre-existing physical or other mental disorders or illicit drug use; and (iii) provision of written informed consent. In total, 71 BD subjects presenting with manic, euthymic, or depressed states were recruited. Among those subjects, 61 subjects were currently undergoing treatment with anti-psychotic medications.
Meanwhile, healthy control (HC) candidates were recruited from the medical examination center of the First Affiliated Hospital at Chongqing Medical University. The inclusion criteria of healthy controls were: (i) no DSM-IV Axis I/Axis II disorder; (ii) no current or previous lifetime history of neurological disease; (iii) no systemic medical illness; and (iv) provision of written informed consents. In total, 126 healthy controls were recruited. Demographic and clinical characteristics of BD subjects and controls were described in table 2.
Table 2 Demographic and clinical characteristics of BD subjects and HCa
Prior to moving forward with the use of potential biomarkers identified in clinical studies, the use of independent samples to validate the biomarkers is essential33. Therefore, the 71 BD subjects and demographically-matched 126 healthy controls were segregated into a training set (~60%) and a test set (~40%). The training set, including 43 BD subjects and 78 demographically-matched healthy controls, was used to identify potential urinary biomarkers and the remaining samples in the test set were used to independently validate the diagnostic performance of these biomarkers.
After overnight fasting, morning urine samples were collected in a sterile cup and transferred into a sterile tube. Urine samples were then centrifuged at 1500 g for 10 min. The resulting supernatant was immediately divided into equal aliquots and stored at −80°C.
NMR acquisition
Prior to NMR analysis, urine samples were thawed and centrifuged at 1500 g for 10 min to remove precipitation. To ensure stabilization of urinary pH, 500 μl of urine was mixed with 100 μl of phosphate buffer (90% D2O, 1 mM 3-trimethylsilyl-1-[2, 2, 3, 3-2H4] propionate (TSP) and 3 mM sodium azide; pH 7.4). After centrifugation at 12000 rpm for 10 min, 500 μl samples of supernatant were transferred into 5 mm NMR tubes. The proton spectra of the urine samples were collected on a Bruker Avance 600 spectrometer operating at a 600.13 MHz 1H frequency with a standard 1-dimensional (1D) pulse sequence. Typically, 64 transients were collected into 16 K data points with a spectral width of 8000 Hz, an acquisition time of 0.945 s and a relaxation delay of 2 s. Prior to Fourier transformation, the free induction decay (FID) was zero-filled and multiplied by an exponential function corresponding to a line-broadening factor of 0.3 Hz in the frequency domain. Urine resonance assignments were performed according to previous literature and NMR databases13,14,34.
GC-MS acquisition
The procedure for GC-MS preparation was performed according to our previous study (15). Briefly, a 15 μl aliquot of urine was vortexed after adding 10 μl internal standard solution (L-leucine-13C6, 0.02 mg/ml). Then, 15 μl urease was added into this mixed solution. The urea was degraded for 60 min at 37°C. The mixture was extracted with 240 μl of ice-cold methanol and then 80 μl of ice-cold methanol. After vortexing for 30 s, the mixture was centrifuged at 14000 rpm for 5 min at 4°C. The 224 ul supernatant was transferred to a glass vial and vacuum-dried at room temperature. The dried metabolic extract was derivatized with 30 μl of methoxyamine (20 mg/ml) for 1.5 h at 37°C. Subsequently, 30 μl of BSTFA with 1% TCMS was added into the mixture and heated for 1 h at 70°C, forming trimethylsilyl (TMS) derivatives. After derivatization and cooling to room temperature, 1.0 μl of this derivative was injected into the GC/MS for analysis. GC/MS analysis was carried out according to this group's previously published work35.
Data analysis
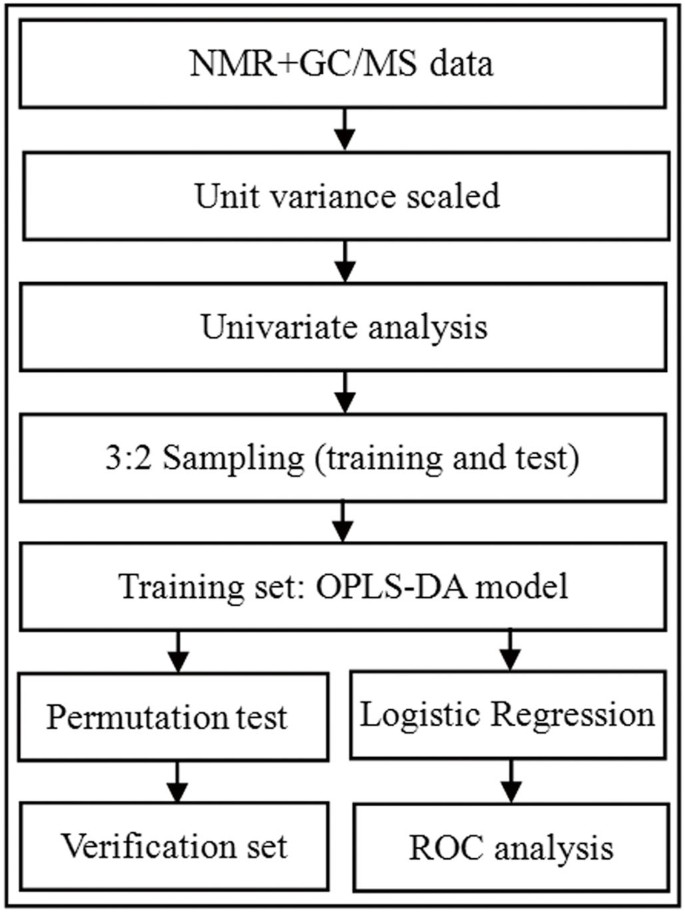
The overall workflow of identifying a simplified set of urinary metabolite biomarkers for BD is summarized in Figure 3. SIMCA-P + 12.0 and SPSS 19.0 were used for all analysis. Spectral data from NMR and GC-MS was collected and unit variance scaled. The data resulting in a _p_-value less than 0.10 in univariate analysis were then used to perform a multivariate analysis25. Orthogonal partial least-squares discriminant analysis (OPLS-DA) was applied to the unit variance-scaled spectral data to visualize discrimination between BD subjects and healthy controls37. Three parameters (R2X, R2Y and Q2Y), calculated by the default leave-one-out (LOO) procedure, were used to describe the quality of OPLS-DA model38. A 99-iteration permutation test was performed to rule out non-randomness of separation between groups. The metabolites with variable importance (VIP) values of no less than 1 (equivalent to a _p_-value of less than 0.5) and a correlation coefficient of |r| > 0.301 (equivalent to a _p_-value of less than 0.5) were identified as the differential metabolites responsible for sample differentiation39,40 and then entered into a multivariate logistic-regression model. A forward selection method was used to obtain a model in which all data had a _p_-value of less than 0.01. In order to make the clinical practice be more feasible and convenient, a step-wise optimization algorithm based on Akaike's information criterion (AIC) was employed to optimize the metabolite biomarker combination41. A receiver-operating characteristic (ROC) curve analysis was used to further evaluate the diagnostic performance of this simplified set of BD biomarkers in the training and test sets. Given the biological reproducibility observed in the independent training and test sets, we repeated the ROC analysis using all subjects to increase the statistical power.
Figure 3
Overview of experimental workflow.
References
- Young, A. H. More good news about the magic ion: lithium may prevent dementia. Br J Psychiatry. 198, 336–7 (2011).
Article Google Scholar - Merikangas, K. R. et al. Prevalence and correlates of bipolar spectrum disorder in the world mental health survey initiative. ARCH GEN PSYCHIAT. 68, 241 (2011).
Article Google Scholar - Müller-Oerlinghausen, B., Berghöfer, A. & Bauer, M. Bipolar disorder. Lancet 359, 241–7 (2002).
Article Google Scholar - Oquendo, M., Currier, D. & Mann, J. Prospective studies of suicidal behavior in major depressive and bipolar disorders: What is the evidence for predictive risk factors? ACTA PSYCHIAT SCAND 114, 151–158 (2006).
Article CAS Google Scholar - Kaddurah-Daouk, R., Kristal, B. S. & Weinshilboum, R. M. Metabolomics: a global biochemical approach to drug response and disease. ANNU REV PHARMACOL 48, 653–83 (2008).
Article CAS Google Scholar - Villas-Boas, S. G. Metabolome Analysis, An Introduction. (Wiley, New Jersey, 2007).
- Weckwerth, W. Metabolomics: Methods and Procedures. (Humana Press, New Jersey, 2007).
- Kristal, B. S. Metabolomics: concept and potential neuroscience application. In Handbook of Neurochemistry and Molecular Neurobiology: Brain Energetics. Integration of Molecular and Cellular Processes. (Springer, New York, 2007).
- Nicholson, J. K. & Lindon, J. C. Systems biology: metabonomics. Nature 455, 1054–1056 (2008).
Article CAS ADS Google Scholar - Jung, J. Y. et al. 1H-NMR-Based Metabolomics Study of Cerebral Infarction. Stroke 42, 1282–8 (2011).
Article CAS Google Scholar - Noga, M. J. et al. Metabolomics of cerebrospinal fluid reveals changes in the central nervous system metabolism in a rat model of multiple sclerosis. Metabolomics 8, 253–263 (2012).
Article CAS Google Scholar - Yang, J. et al. Potential metabolite markers of schizophrenia. Mol Psychiatry. 18, 67–78 (2011).
Article Google Scholar - Yap, I. K. et al. Nicholson. Urinary metabolic phenotyping differentiates children with autism from their unaffected siblings and age-matched controls. J PROTEOME RES. 9, 2996–3004 (2010).
Article CAS Google Scholar - Zheng, P. et al. Identification and validation of urinary metabolite biomarkers for major depressive disorder. Mol Cell Proteomics. 12, 207–14 (2013).
Article Google Scholar - Zheng, P. et al. A novel urinary metabolite signature for diagnosing major depressive disorder. J Proteome Res. 12, 5904–11 (2013).
Article CAS ADS Google Scholar - Zheng, P. et al. Plasma metabonomics as a novel diagnostic approach for major depressive disorder. J Proteome Res. 11, 1741–8 (2012).
Article CAS Google Scholar - Lan, M. et al. Metabonomic analysis identifies molecular changes associated with the pathophysiology and drug treatment of bipolar disorder. MOL PSYCHIATR. 14, 269–279 (2008).
Article Google Scholar - Sussulini, A. et al. Metabolic profiling of human blood serum from treated patients with bipolar disorder employing 1H NMR spectroscopy and chemometrics. ANAL CHEM 81, 9755–63(2009).
Article CAS Google Scholar - Zheng, P. et al. Novel urinary biomarkers for diagnosing bipolar disorder. Metabolomics. 9, 800–808 (2013).
Article CAS Google Scholar - Xu, X. J. et al. 2, 4-Dihydroxypyrimidine is a potential urinary metabolite biomarker for diagnosing bipolar disorder. Mol Biosyst. 10, 813–9 (2014).
Article CAS Google Scholar - Williams, R. et al. A multi-analytical platform approach to the metabonomic analysis of plasma from normal and Zucker (fa/fa) obese rats. Mol Biosyst. 2, 174–83 (2006).
Article CAS Google Scholar - Bouatra, S. et al. The human urine metabolome. PLoS One 8, e73076 (2013).
Article CAS ADS Google Scholar - Law, W. S. et al. Metabonomics investigation of human urine after ingestion of green tea with gas chromatography/mass spectrometry, liquid chromatography/mass spectrometry and H-1 NMR spectroscopy. Rapid Commun Mass Spectrom 22, 2436–2446 (2008).
Article CAS ADS Google Scholar - Li, Y. et al. Metabonomics study of essential hypertension and its chinese medicine subtypes by using gas chromatography-mass spectrometry and nuclear magnetic resonance spectroscopy. Evid Based Complement Alternat Med. 2013, 625906 (2013).
ADS PubMed PubMed Central Google Scholar - Andreazza, A. C. et al. Oxidative stress markers in bipolar disorder: a meta-analysis. J AFFECT DISORDERS. 111, 135–144 (2008).
Article CAS Google Scholar - Löffler, M., Fairbanks, L. D., Zameitat, E., Marinaki, A. M. & Simmonds, H. A. Pyrimidine pathways in health and disease. Trends Mol Med. 11, 430–7 (2005).
Article Google Scholar - Xia, J. G., Mandal, R., Sinelnikov, I. V., Broadhurst, D. & Wishart, D. S. MetaboAnalyst 2.0-a comprehensive server for metabolomic data analysis. NUCLEIC ACIDS RES. 40, W127–33 (2012).
Article CAS Google Scholar - Micheli, V. et al. Neurological disorders of purine and pyrimidine metabolism. Curr Top Med Chem. 11, 923–47 (2011).
Article CAS Google Scholar - Chu, T. T., Liu, Y. & Kemether, E. Thalamic transcriptome screening in three psychiatric states. J Hum Genet. 54, 665–75 (2009).
Article CAS Google Scholar - Wu, F. S., Gibbs, T. T. & Farb, D. H. Dual activation of GABAA and glycine receptors by beta-alanine: inverse modulation by progesterone and 5 alpha-pregnan-3 alpha-ol-20-one. Eur J Pharmacol. 246, 239–46 (1993).
Article CAS Google Scholar - Fatemi, S. H., Folsom, T. D., Rooney, R. J. & Thuras, P. D. Expression of GABAA α2-, β1- and ε-receptors are altered significantly in the lateral cerebellum of subjects with schizophrenia, major depression and bipolar disorder. Transl Psychiatry. 3, e303 (2013).
Article CAS Google Scholar - Zarate, C. A., Singh, J. & Manji, H. K. (2006) Cellular plasticity cascades: targets for the development of novel therapeutics for bipolar disorder.Biol Psychiatry. 59, 1006–1020 (2006).
Article CAS Google Scholar - Micheel, C. Evolution of Translational omics: Lessons Learned and the Path Forward. (Institute of Medicine Consensus Report). (National Academies Press, Washington, 2012).
- Beckwith-Hall, B. et al. Nuclear magnetic resonance spectroscopic and principal components analysis investigations into biochemical effects of three model hepatotoxins. CHEM RES TOXICOL. 11, 260–272 (1998).
Article CAS Google Scholar - Shao, W. H. et al. Metabolomic identification of molecular changes associated with stress resilience in the chronic mild stress rat model of depression. Metabolomics. 9, 433–443 (2012).
Article Google Scholar - Roubaud-Baudron, C., Krolak-Salmon, P., Quadrio, I., Mégraud, F. & Salles, N. Impact of chronic Helicobacter pylori infection on Alzheimer's disease: preliminary results. Neurobiol Aging. 33, 1009.e11–9 (2012).
Article CAS Google Scholar - Bylesj, M. et al. OPLS discriminant analysis: combining the strengths of PLS-DA and SIMCA classification. J Chemom. 20, 341–351 (2006).
Article Google Scholar - Mahadevan, S., Shah, S. L., Marrie, T. J. & Slupsky, C. M. Analysis of metabolomic data using support vector machines. Anal Chem. 80, 7562–7570 (2008).
Article CAS Google Scholar - Cloarec, O. et al. Evaluation of the orthogonal projection on latent structure model limitations caused by chemical shift variability and improved visualization of biomarker changes in 1H NMR spectroscopic metabonomic studies. Anal Chem. 77, 517–526 (2005).
Article CAS Google Scholar - Qi, Y. et al. Urinary metabolite markers of precocious puberty. Mol Cell Proteomics. 11, M111.011072 (2012).
Article Google Scholar - Oikonomopoulou, K. et al. Prediction of ovarian cancer prognosis and response to chemotherapy by a serum-based multiparametric biomarker panel. Br J Cancer. 99, 1103–1113 (2008).
Article CAS Google Scholar
Acknowledgements
Our sincere gratitude is extended to Professors Delan Yang and Hua Hu for their efforts in sample collection. We also thank Dr. N.D. Melgiri for editing and proofreading the manuscript. This work was supported by the National Youth Division (31300881), the Postdoctoral Fund (XM201101003) and the National Basic Research Program of China (973 Program, grant no. 2009CB918300).
Author information
Author notes
- Chen Jian-jun, Liu Zhao, Fan Song-hua and Yang De-yu contributed equally to this work.
Authors and Affiliations
- Department of Neurology, The First Affiliated Hospital, Chongqing Medical University, Chongqing, China
Jian-jun Chen, Zhao Liu, Song-hua Fan, Peng Zheng, Wei-hua Shao, Xue-jiao Xu, Qi Li, Jun Mu, Yong-tao Yang & Peng Xie - Institute of Neuroscience, Chongqing Medical University, Chongqing, China
Jian-jun Chen, Zhao Liu, Song-hua Fan, De-yu Yang, Peng Zheng, Wei-hua Shao, Zhi-guo Qi, Xue-jiao Xu, Qi Li, Jun Mu, Yong-tao Yang & Peng Xie - Chongqing Key Laboratory of Neurobiology, Chongqing Medical University, Chongqing, China
Jian-jun Chen, Zhao Liu, Song-hua Fan, De-yu Yang, Peng Zheng, Wei-hua Shao, Zhi-guo Qi, Xue-jiao Xu, Qi Li, Jun Mu, Yong-tao Yang & Peng Xie
Authors
- Jian-jun Chen
You can also search for this author inPubMed Google Scholar - Zhao Liu
You can also search for this author inPubMed Google Scholar - Song-hua Fan
You can also search for this author inPubMed Google Scholar - De-yu Yang
You can also search for this author inPubMed Google Scholar - Peng Zheng
You can also search for this author inPubMed Google Scholar - Wei-hua Shao
You can also search for this author inPubMed Google Scholar - Zhi-guo Qi
You can also search for this author inPubMed Google Scholar - Xue-jiao Xu
You can also search for this author inPubMed Google Scholar - Qi Li
You can also search for this author inPubMed Google Scholar - Jun Mu
You can also search for this author inPubMed Google Scholar - Yong-tao Yang
You can also search for this author inPubMed Google Scholar - Peng Xie
You can also search for this author inPubMed Google Scholar
Contributions
Conceived and designed the experiments: P.X. J.J.C. Z.L. and S.H.F. Performed the experiments: D.Y.Y. P.Z. W.H.S. and Y.T.Y. Analyzed the data: J.J.C. Z.L. and Z.G.Q. Contributed reagents/materials/analysis tools: X.J.X. Q.L. and J.M. Wrote the paper: J.J.C. Z.L. S.H.F. and D.Y.Y.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Chen, Jj., Liu, Z., Fan, Sh. et al. Combined Application of NMR- and GC-MS-Based Metabonomics Yields a Superior Urinary Biomarker Panel for Bipolar Disorder.Sci Rep 4, 5855 (2014). https://doi.org/10.1038/srep05855
- Received: 08 May 2014
- Accepted: 10 July 2014
- Published: 28 July 2014
- DOI: https://doi.org/10.1038/srep05855