The brave new world of RNA (original) (raw)
Most of the RNA transcribed from your genome doesn't make protein. Carina Dennis talks to the revolutionaries who believe that it functions in gene-regulatory networks that underlie the complexity of higher organisms.

Denying dogma: John Mattick believes that non-coding RNAs underpin complex gene regulation.
Biology's 'central dogma', laid down in the 1950s, states that genetic information flows from DNA to RNA to protein. Since then, numerous studies have shown that RNA does more than simply serve this intermediary function. But is it time to cast dogma aside and completely rethink the role of RNA? The answer is yes, according to a handful of geneticists. In multicellular organisms, they argue, the majority of RNA molecules are the principal actors in largely unexplored networks of gene regulation.
What's more, these researchers suggest that understanding RNA-based gene-regulatory networks may provide the key to explaining the difference between a yeast cell and a fruitfly, and between a fruitfly and you or me. “Complexity is hidden in the non-coding output of the genome,” asserts John Mattick of the University of Queensland in Brisbane, Australia, who has published a series of papers that form an expansive but still speculative theoretical backdrop to the emerging view of RNA as a regulator of gene activity1,2,3.
Many experts remain unconvinced. But as research in genomics reveals a vast array of RNAs that do not code for proteins, many biologists are finding it difficult to believe that they have no function. And with the discovery of a new class of non-coding RNAs, dubbed microRNAs (miRNAs), that seem to regulate the production of proteins from other genes, Mattick's ideas don't seem quite so outlandish.
In bacteria, most DNA codes for proteins. Geneticists have long known that the genomes of higher organisms are littered with non-coding DNA, much of which is never read at all. But some of it is transcribed to RNA — your genes, for instance, contain non-coding 'introns' between the 'exons' that contain the instructions for making proteins. When a messenger RNA (mRNA) is transcribed from a gene, the introns are cut out and the exons spliced together. Many sequences outside protein-coding genes are also transcribed into RNA.
Mattick claims that introns and other non-coding RNAs may account for up to 98% of the transcriptional output of the human genome2. He argues that non-coding RNAs interact with one another, with mRNAs, with DNA and with proteins to form networks that can regulate gene activity with almost infinite potential complexity.
It's an appealing idea, because comparisons of gene numbers don't seem to explain the difference between simple and complex organisms. We have only about two or three times as many protein-coding genes as the nematode Caenorhabditis elegans or the fruitfly Drosophila melanogaster, which, in turn, have only about twice as many as the yeast Saccharomyces cerevisiae. Higher organisms seem to bolster their complexity by mixing and matching protein domains to generate new combinations. But other ploys may be needed to account for the vast complexity of humans and other vertebrates.
Mattick likes to explain the workings of his proposed RNA-based networks in terms of computer science. The traditional biological view, based on the central dogma, is like a computer that would have to be rewired to perform each new calculation. An RNA network that interacts with all levels of the path from DNA to protein, however, could allow many different outputs to be derived from the same basic genetic circuitry, just as a computer's controlling software allows its processor to be easily reconfigured for a new task by changing the control codes. “It is not surprising that, for the evolution of complex systems, changes came about primarily in the control architecture rather than the components,” Mattick argues.
Transcript trawl
Accumulating the evidence to confirm or disprove this theory may take years. But analyses of the human genome are already showing that the realm of non-coding RNA is much bigger than most biologists had realized. In May, a team led by Thomas Gingeras, vice president for biological research at Affymetrix in Santa Clara, California, used DNA microarrays to probe each stretch of sequence across human chromosomes 21 and 22 for evidence of transcription. The researchers found that much more DNA was being transcribed than expected, given the number of genes that are thought to be present on these chromosomes4. “We saw ten times more sequence being transcribed than was predicted,” says Gingeras.
Just showing that non-coding RNAs are common isn't enough to prove that most are involved in networks of gene regulation. But the discovery of miRNAs has given some credence to the idea. At roughly 22 nucleotides long, miRNAs passed unnoticed for a long time. The first genes coding for miRNAs — called lin-4 and let-7 — were identified in C. elegans in 1993 and 2000, respectively5,6,7. The miRNAs are cut from longer, hairpin-shaped RNAs transcribed from lin-4 and let-7, and bind to specific target mRNAs, blocking their translation to proteins.

Feline contrary: Sean Eddy contends that many non-coding RNAs are just “transcriptional slop”.
This new class of RNAs was initially considered to be peculiar to nematodes, until a team led by Gary Ruvkun of Harvard Medical School in Boston found that let-7 is present in a diverse range of species, from vertebrates, including humans and zebrafish, to fruitflies and molluscs8.
The tally of miRNAs continues to increase. “It feels like a snowball about to turn into an avalanche,” says Lawrence Hurst, an evolutionary geneticist at the University of Bath, UK. More than 150 miRNAs have now been identified from various animals, as a handful of groups have begun to prospect for them in earnest9,10,11,12,13. And this year has seen the first reports of their presence in plants14,15.
Just how many miRNAs exist is not yet known. “We are still developing the technology to identify miRNAs, so it is difficult to make an estimate,” says Victor Ambros of Dartmouth Medical School in Hanover, New Hampshire, who led one of the teams that first identified lin-4.
At the University of Münster in Germany, for instance, researchers led by Alexander Hüttenhofer are studying libraries of expressed sequences from mouse brain tissue. Whereas researchers hunting for protein-coding genes typically look for mRNAs in such extracts using methods that recognize characteristic structures called 'poly-A' tails, Hüttenhofer's team abandons this approach and uses gel electrophoresis to separate out smaller RNAs. “People looking for genes isolated extracts enriched for mRNA and threw away the remainder. We are the garbage people of the human genome project,” says Hüttenhofer. So far, his team's search through the trash has yielded hundreds of small, non-coding RNAs16 — at least some of which are likely to be miRNAs.
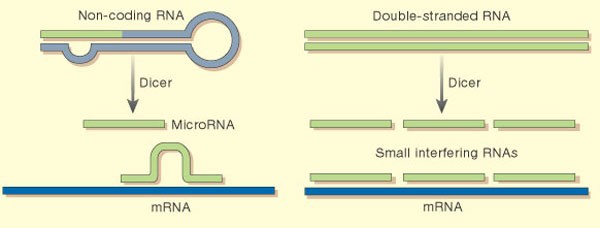
The way in which miRNAs are generated has revealed an intriguing link to a gene-silencing mechanism known as RNA interference (RNAi), which is thought to defend cells from viruses and 'jumping genes'17. It springs into action when the cell detects an unusual RNA with paired strands. An enzyme called Dicer cuts up the offending double-stranded RNA, creating fragments 21–25 nucleotides long, called small interfering RNAs (siRNAs). Single strands from these fragments bind to further copies of the original RNA, targeting them for destruction. RNAi can also be used experimentally to silence a cell's own genes, by adding double-stranded RNA sequences that match a gene's mRNA.
Making the cut
Studies of lin-4 and let-7 have revealed that Dicer cuts the miRNAs from the two genes' transcripts18,19. They then bind to their target mRNA sequences, albeit less perfectly than the binding between siRNAs and their targets, causing them to bulge out (see diagram). The end result is also different: miRNAs merely prevent mRNAs from making proteins; they do not cause them to be destroyed. “My hunch is that RNAi is the ancestral pathway,” says Ambros, who believes that aspects of its mechanism were subsequently co-opted to regulate the cell's own genes.
Figure 1
The enzyme Dicer generates microRNAs that bind imperfectly to target mRNAs, blocking protein production (left); in RNAi, the RNAs made by Dicer bind tightly to mRNAs, targeting them for doom.

Phillip Zamore says that it would be a cruel joke if conserved microRNAs have no function. Credit: UMASS MED. SCHOOL
Because miRNAs do not destroy their targets, it is possible that they may impose a temporary halt on the translation of mRNA to protein that can be relieved on cue. “Suppressing translation rather than destroying the transcripts seems a better way to modulate gene expression, as it leaves open the potential for switching translation back on again,” says Thomas Tuschl of the Max Planck Institute for Biophysical Chemistry in Göttingen, Germany.
The fact that the sequences of the miRNAs discovered so far are remarkably similar across different species suggests that most may share the gene-regulatory role played by lin-4 and let-7. “It would be a cruel joke that nature has played on scientists if the sequences had been conserved in evolution but with no function,” says Phillip Zamore of the University of Massachusetts Medical School in Worcester.
Showing that most miRNAs have real biological significance, however, will require systematic attempts to see what happens if they are mutated — something that is easier said than done. “miRNAs have largely been missed by genetic screens because they are such small targets for mutagenesis,” says David Bartel of the Whitehead Institute for Biomedical Research in Cambridge, Massachusetts, who is now collaborating in a project to systematically create miRNA mutants in C. elegans.

Small target: David Bartel says genetic screens have missed microRNAs because of their size. Credit: K. DOOHER
Nevertheless, the limited evidence available supports the idea that miRNAs are involved in gene regulation. Some fruitfly mRNAs, for instance, contain sequence motifs that allow translation to the corresponding proteins to be shut down when required. And Eric Lai of the University of California, Berkeley, has found that some miRNAs have sequences that would bind perfectly to these motifs, suggesting that they may be switches that turn off protein production20.
miRNAs are not the only non-coding RNA sequences that have been shown to play a role in regulating gene activity. A gene called XIST, for instance, encodes a large RNA transcript that shuts down the 'spare' X chromosome in female mammals21. And in mice, a non-coding RNA dubbed Air has recently been shown to be involved in a gene-silencing mechanism that can shut down one of the two copies of a gene that are inherited, one from each parent22. Mattick is convinced that a wealth of further regulatory RNAs remains to be discovered. “This is just the tip of the iceberg,” he says.
But even among researchers studying non-coding RNAs, many believe that their gene-regulatory role could be relatively limited. Although many miRNAs may remain to be discovered, for now they are vastly outnumbered by the tens of thousands of known protein-coding genes. And as for the rest of the vast, uncharted landscape of non-coding RNA, it could still largely be garbage, after all.
Messy business
Many researchers believe that the process of transcribing RNA from DNA is inherently messy. “My opinion is simply that transcription might be a noisy process, and that a lot of RNAs are made for no good reason,” says Jean-Michel Claverie of the Institute of Structural Biology and Microbiology in Marseille, part of the CNRS, France's national research agency. Sean Eddy, a computational biologist at Washington University in St Louis, Missouri, suspects that the cell rationalizes the costs of running a watertight operation against allowing some leakage — and leaky transcription wins out. “It's cheap to make transcripts,” he argues. “The cell can tolerate a high level of transcriptional slop.”
But Hurst disagrees. “Many developmental pathways require exquisite control of gene expression,” he observes. “There are many reasons why a leaky genome is not desired.” And that view receives support from unpublished studies by Gingeras, who has compared the DNA sequences transcribed to non-coding RNA from human chromosomes 21 and 22 to the corresponding regions of the mouse and zebrafish genomes. The sequences are very similar, suggesting that they have been conserved by evolution because they perform some useful function.
Although the theory of RNA-based gene regulation seems attractive, putting it to the test won't be easy. Building on Gingeras's observations, one idea is to begin by looking for non-coding sequences that are conserved between the genomes of related species and determining if they are transcribed into RNA, before attempting the painstaking bench work needed to investigate the transcripts' function.
Eddy and others are also trying to devise computational strategies that can identify genes for functional non-coding RNAs from genome sequence data. Many of the non-coding RNAs that have so far been shown to have definite functions fold up into characteristic 'secondary' structures — so being able to predict these structures from DNA sequence data may help in the search for new non-coding RNAs.
Mattick, meanwhile, is looking for sequences within introns in yeast that match those in other protein-coding genes, reasoning that the former may produce RNAs that interact with the latter. In parallel, his team is conducting computer simulations of hypothetical RNA-based gene-regulatory networks, to see whether it is possible to generate immense complexity from simple beginnings simply by altering the regulatory network.

Go to page 213 for our Insight on RNA
Potentially, gene regulation by non-coding RNA could not only provide the solution to the mystery of how higher organisms have generated such mind-boggling complexity from a relatively limited genetic palette, but may also explain much of the variation between individuals within a population — including the occurrence of some diseases. Intriguingly, Tuschl's team has mapped the genetic aberration that causes chronic lymphoid leukaemia to a region of the genome that includes a gene encoding an miRNA, making it a plausible candidate.
But even if non-coding RNA turns out not to have overriding biological significance, the genome project's garbage people have already sifted some gems from the trash. “Five or six years ago, people said we were wasting our time,” says Hüttenhofer. Today no one regards people studying non-coding RNA as time-wasters.