A nonsense mutation in FMR1 causing fragile X syndrome (original) (raw)
Introduction
Fragile X syndrome is a frequent form of inherited intellectual disability affecting approximately 1 in 4000 males (reviewed by Chonchaiya et al.1). In the vast majority of patients the syndrome is caused by expansion of a CGG repeat located in the 5′UTR of the FMR1 gene localized to Xq27.3. The gene was cloned in 1991 and comprises 17 coding exons. The protein, FMRP, is an RNA-binding protein with 632 amino acids.2 Only four point mutations in FMR1 have been reported: a missense mutation p.Ile304Asn, a 1-bp deletion c.373delA in exon 5 resulting in a frameshift and premature truncation of the protein, a 2-bp change g.23714GG>TA spanning the intron/exon boundary of exon 2, and a missense mutation p.Arg138Gln.3, 4, 5 Deletions of the CGG repeat and flanking sequences have been reported several times.6
Patients and methods
Patient
The patient, a 35-year-old mentally retarded man, was referred to a tertiary epilepsy centre due to difficulties in treatment of epilepsy, with absences, generalised tonic–clonic seizures and myoclonias. On further diagnostic evaluation he was tested for fragile X syndrome. Phenotypically he had classical fragile X syndrome, with an elongated face, high, broad forehead, low-set large ears, prognathia and enlarged testes. Neurological examination showed hypotonia and hypermobility, with hyperextensible joints. He had no active language except for a few repeated words used out of context, and showed autistic features with little eye contact, discomfort upon physical contact, perseveration and limited interests. He was described by his mother as showing late motor development in childhood and having need for speech therapy, and special day-care and schooling. Epilepsy debuted at age 4 years.
The mother presented with mild-to-moderate intellectual disability. She reported special education needs and carried out sheltered work. She was married and capable of maintaining a household. Physical examination showed a relatively unremarkable phenotype, but her behaviour included hypermotor activity and many automatisms. She was capable of a simple conversation.
Molecular biology procedures
Genomic DNA was extracted from peripheral blood lymphocytes (EDTA stabilized) using standard procedures. Lymphoblastoid cell lines were established from the patient and his mother using Epstein–Barr virus transformation.
Southern blot analysis was performed as described.7 Briefly, 9 μg of high molecular genomic DNA was digested with _Pst_1, and 6 μg DNA was digested with _Eco_R1 plus _Eag_I and size-separated on an agarose gel. Standard blotting technique was used to transfer DNA to a nitrocellulose filter, which was hybridized with radioactive labelled pPX6 probe. After washing, the filter was analysed using a Cyclone from Perkin-Elmer (Waltham, MA, USA).
PCR analysis of the FMR1 CGG repeat was performed using the fragile X kit from Abbott Molecular (IL, USA) following the manufacturer's instructions. Western blot analysis was performed as described.7
Mutational analysis of FMR1 was performed by direct DNA sequencing of PCR products of coding exons and at least 20 bp of flanking sequences. Primers and PCR conditions are available upon request. Sequencing was performed using BigDye v3.1 terminator chemistry and an ABI3130XL genetic analyzer and using the Seqscape program for analysis (Applied Biosystems, Foster City, CA, USA). X-inactivation analysis was performed using the polymorphic CAG repeat in the androgen receptor locus as described.7
Results
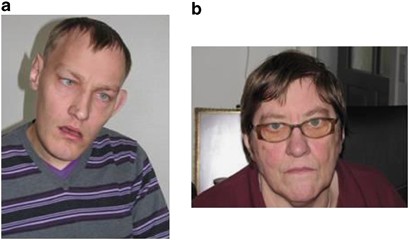
The patient and his mother are shown in Figure 1.
Figure 1
Facial appearance of (a) the patient and (b) his mother. Note the elongated face, high broad forehead and prognathia in the patient (a).
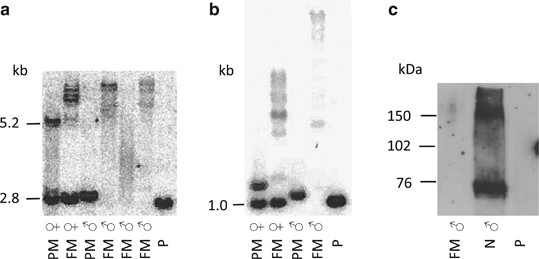
Southern blot analyses of FRAXA (_Eco_RI+_Eag_I and _Pst_I digestion, respectively) showed band patterns corresponding to a repeat number in the normal range (Figures 2a and b). PCR analysis showed an allele size corresponding to 29 repeats. However, based on the classic clinical fragile X appearance, we performed a western blot using an anti-FMRP antibody, which showed that no FMRP was expressed (Figure 2c and Supplementary Figure 1). We also performed Southern blot analysis and PCR of FRAXA of the mother of the patient; this showed a normal band pattern and repeat numbers of 29 and 33, with no indication of a full mutation (data not shown). Subsequent DNA sequencing revealed a sequence variation in exon 2 of FMR1 c.80C>A resulting in a nonsense mutation p.Ser27X (Supplementary Figure 2). Verification of the sequence variation was performed on a second PCR product, and the variation was seen in both directions. The mother was found to be heterozygous for the mutation. We investigated X-inactivation pattern using a polymorphic CAG repeat in the androgen receptor locus and found an equal distribution of active and inactive X-chromosomes in blood (data not shown).
Figure 2
Southern blot analysis using (a) _Eco_RI+_Eag_I digestion and (b) _Pst_I digestion. P, patient; M, mother; FM ♀, full mutated female; PM ♀, premutated female; FM ♂, full mutated male; PM ♂, premutated male; N ♂, normal male; N ♀, normal female. (c) Western blot analysis using an anti-FMRP antibody. P, patient; FM ♂, full mutated male; N ♂ normal male.
Discussion
We report a nonsense mutation in FMR1 in a patient with classic fragile X syndrome and his carrier mother with mild intellectual impairment. The fragile X syndrome can be considered semi-dominant, with manifestations of a full mutation in female carriers, depending on the proportion of active versus inactive X chromosomes carrying the mutation. Earlier studies have shown that phenotypical normal females have a skewed X inactivation pattern in favour of the inactive X carrying the mutation.8 As the X inactivation in the mother showed an equal distribution, some degree of intellectual impairment would be expected. However, this study was performed in blood and therefore is an approximation, as we do not know the pattern in brain.
Surprisingly, few point mutations have been found in the FMR1 gene. Two of these are truncating mutations in the N-terminal half of the gene and supposedly lead to nonsense-mediated decay of the mRNA. These two mutations caused classical fragile X syndrome.5 Another patient had a missense mutation in one of the KH domains and had a more severe phenotype.4 This could be due to a gain-of-function effect; however, in this family X-linked glycogenosis was also present, which might have contributed to the phenotype. The observed low frequency of point mutations in FMR1 might be because FMR1 is not routinely screened for point mutations, but is only investigated for the common CGG expansion; another reason could be that point mutations in this gene give atypical or even lethal phenotypes. More than 1100 patients have been screened for point mutations in FMR1;3, 9, 10, 11, 12, 13, 14, 15 however, only one missense mutation of questionable pathogeneity was identified.3 The individuals investigated in these studies represent a very heterogenous group of patients with developmental delay, indicating that FMR1 mutations are not a common cause for mental impairment. To our knowledge, a mutational screening of FMR1 has not been performed in a large cohort of patients with typical fragile X syndrome negative for repeat expansion. Therefore, the frequency of point mutations in FMR1 in classical fragile X syndrome is unknown. The patient presented in this study shows a typical fragile X phenotype, and his mother shows a phenotype comparable to a female with a full mutation. We therefore suggest that in patients with clinical fragile X syndrome and no CGG expansion expanded molecular diagnosis should be considered, including western blot analysis and DNA sequencing.
References
- Chonchaiya W, Schneider A, Hagerman RJ : Fragile X: a family of disorders. Adv Pediatr 2009; 56: 165–186.
Article Google Scholar - Verkerk AJ, Pieretti M, Sutcliffe JS et al: Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 1991; 65: 905–914.
Article CAS Google Scholar - Collins SC, Bray SM, Suhl JA et al: Identification of novel FMR1 variants by massively parallel sequencing in developmentally delayed males. Am J Med Genet A 2010; 10: 2512–2520.
Article Google Scholar - DeBoulle K, Verkerk AJMH, Reyniers E et al: A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat Genet 1993; 3: 31–35.
Article CAS Google Scholar - Lugenbeel KA, Peier AM, Carson NL, Chudley AE, Nelson DL : Intragenic loss of function mutations demonstrate the primary role of FMR1 in fragile X syndrome. Nat Genet 1995; 10: 483–485.
Article CAS Google Scholar - Coffee B, Ikeda M, Budimirovic DB, Hjelm LN, Kaufmann WE, Warren ST : Mosaic FMR1 deletion causes fragile X syndrome and can lead to molecular misdiagnosis: a case report and review of the literature. Am J Med Genet A 2008; 146A: 1358–1367.
Article CAS Google Scholar - Gronskov K, Hjalgrim H, Bjerager MO, Brondum-Nielsen K : Deletion of all CGG repeats plus flanking sequences in FMR1 does not abolish gene expression. Am J Hum Genet 1997; 61: 961–967.
Article CAS Google Scholar - Martinez R, Bonilla-Henao V, Jimenez A et al: Skewed X inactivation of the normal allele in fully mutated female carriers determines the levels of FMRP in blood and the fragile X phenotype. Mol Diagn 2005; 9: 157–162.
Article Google Scholar - Castellvi-Bel S, Sanchez A, Badenas C et al: Single-strand conformation polymorphism analysis in the FMR 1 gene. Am J Med Genet 1999; 84: 262–265.
Article CAS Google Scholar - Chiurazzi P, de Graff E, Ng J et al: No apparent involvement of the FMR1 gene in five patients with phenotypic manifestations of the fragile X syndrome. Am J Med Genet 1994; 51: 309–314.
Article CAS Google Scholar - Collins SC, Coffee B, Benke PJ et al: Array-based FMR1 sequencing and deletion analysis in patients with a fragile X syndrome-like phenotype. PLoS One 2010; 5: e9476.
Article Google Scholar - Gronskov K, Hallberg A, Brondum-Nielsen K : Mutational analysis of the FMR1 gene in 118 mentally retarded males suspected of fragile X syndrome: absence of prevalent mutations. Hum Genet 1998; 102: 440–445.
Article CAS Google Scholar - Reyniers E, Wolff G, Tariverdian G et al: Severe mental retardation and macroorchidism without mutation in the FMR1 gene. Am J Med Genet 1996; 64: 408–412.
Article CAS Google Scholar - Shinahara K, Saijo T, Mori K, Kuroda Y : Single-strand conformation polymorphism analysis of the FMR1 gene in autistic and mentally retarded children in Japan. J Med Invest 2004; 51: 52–58.
Article Google Scholar - Wang YC, Lin ML, Lin SJ, Li YC, Li SY : Novel point mutation within intron 10 of FMR-1 gene causing fragile X syndrome. Hum Mutat 1997; 10: 393–399.
Article CAS Google Scholar
Author information
Authors and Affiliations
- Center for Applied Human Molecular Genetics, The Kennedy Center, Glostrup, Denmark
Karen Grønskov, Karen Brøndum-Nielsen, Alma Dedic & Helle Hjalgrim - Genetic Counseling Clinic, The Kennedy Center, Glostrup, Denmark
Karen Brøndum-Nielsen - Danish Epilepsy Centre, Dianalund, Denmark
Helle Hjalgrim
Authors
- Karen Grønskov
You can also search for this author inPubMed Google Scholar - Karen Brøndum-Nielsen
You can also search for this author inPubMed Google Scholar - Alma Dedic
You can also search for this author inPubMed Google Scholar - Helle Hjalgrim
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toKaren Grønskov.
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Grønskov, K., Brøndum-Nielsen, K., Dedic, A. et al. A nonsense mutation in FMR1 causing fragile X syndrome.Eur J Hum Genet 19, 489–491 (2011). https://doi.org/10.1038/ejhg.2010.223
- Received: 20 September 2010
- Revised: 15 November 2010
- Accepted: 16 November 2010
- Published: 26 January 2011
- Issue Date: April 2011
- DOI: https://doi.org/10.1038/ejhg.2010.223