Gene–environmental interaction regarding alcohol-metabolizing enzymes in the Japanese general population (original) (raw)
Introduction
Restraint of drinking (or alcohol consumption) is often recognized as an important strategy for lifestyle modification to prevent people from developing hypertension and cardiovascular diseases. This is because chronic alcohol consumption has been shown to moderately correlate with these disorders.1, 2 However, the degree of correlation is known to vary among individuals.
It has been known that there exist racial differences in alcohol metabolism that appear to be substantially influenced by genetic factors. For example, the proportion of people with low alcohol-metabolizing activity is greater in Asians than in Caucasians.3 Moreover, it has been reported that gene polymorphisms in some alcohol-metabolizing enzymes significantly influence the risk of developing alcoholism, that is, addictive status of drinking.4 In the general population, drinking behavior (or alcohol preference) is supposed to be principally defined by a combination of factors including genetic and environmental factors. The genetic factors regulating drinking behavior may also determine the sensitivity to the pressor effects of alcohol.
Thus far, only a few studies have systematically investigated in the population-based setting the relationship between drinking behavior and gene polymorphisms in the alcohol-metabolizing enzymes. Moreover, earlier studies have not fully examined the relationship between genetic factors regulating drinking behavior and blood pressure elevation (or hypertension) induced by chronic alcohol consumption.
Under these circumstances, toward the establishment of a scientific basis for encouraging people in sustained abstinence or restraint of alcohol consumption, this study was designed (1) to perform candidate gene approach by exploring a list of gene polymorphisms that could determine individual drinking behavior, (2) to evaluate their genetic impacts in the large-scale Japanese general population and (3) to investigate the influence of genetic susceptibility to drinking behavior on the sensitivity to the pressor effects of alcohol.
Methods
Study subjects
In this study, we consecutively enrolled individuals who sought medical assessment from September 2002 to August 2003 at the Amagasaki Health Medical Foundation, Hyogo prefecture. Participants were included when they were over 18 years of age and had full clinical examination data, along with a completed questionnaire on lifestyle. On the basis of these inclusion criteria, 5451 individuals that comprised 3296 male participants and 2155 female participants were enrolled in the study. During the medical assessment, the height and weight of the subjects were measured and the body mass index (BMI) calculated; blood pressure was measured using an automatic sphygmomanometer in the sitting position after at least 5 min of rest. For the purpose of quantitative analysis, we adopted cross-sectional measurements uniformly taken from each subject, whereas people taking antihypertensive treatment were excluded. All study subjects were not blood relatives but provided their written informed consent for participation. The protocol of the study was approved by the Ethics Committee of the International Medical Center of Japan.
Assessment of lifestyle
Drinking and smoking behavior was assessed on the basis of the lifestyle questionnaire self-administered by the subjects. According to the frequency of drinking, the subjects were categorized into three groups: nondrinker (0 time in a week), chance drinker (less than four times in a week) and regular drinker (equal to or more than four times in a week). For chance and regular drinkers, alcohol intake was reported as the usual amounts that were denoted in terms of servings of sake (1 gou [180 ml] of Japanese rice wine is considered equal to 22 g of ethanol). According to the amounts of alcohol consumed, the regular drinkers were further classified into two groups: subjects who consumed less than 1 gou day−1 (<1 gou day−1) and those who consumed equal to or greater than 1 gou day−1 (⩾1 gou day−1). Also, according to the smoking habits, subjects were classified into three groups: nonsmokers, past smokers and current smokers.
Detection of tag SNPs in four candidate genes
In alcohol metabolism, alcohol dehydrogenase (ADH) oxidizes ethanol in alcoholic beverages to acetaldehyde, and aldehyde dehydrogenase (ALDH) converts it to acetic acid. Both the enzymes play a key role in alcohol metabolism. Thus, we chose four genes, ADH1A, ADH1B, ADH1C and ALDH2, as candidate genes. Potential functional single nucleotide polymorphisms (SNPs) were screened by resequencing of genomic DNA derived from 48 Japanese subjects in all exons, 5′-untranslated regions (UTRs) and 3′-UTRs of each gene (Supplementary Table S1A and B). The information of identified SNPs was deposited in the NCBI's SNP database (http://www.ncbi.nlm.nih.gov/SNP/) and also in our own database, JMDBase (http://www.jmdbase.jp/JmdBaseExt/Top.aspx).
Tag SNPs were selected for four genes with the algorithm that we reported earlier.5 Briefly, by categorizing any pair of SNPs having linkage disequilibrium (LD) coefficient _r_2⩾0.6 into the same LD group, we defined LD block and selected tag SNPs from each LD group. These tag SNPs were then used for genotyping assay. LD relations between SNPs in four target genes were graphically shown as LD plots (Supplementary Figure S1). The information on the LD group and selected tag SNPs is shown in Supplementary Table S1A and B.
Genotyping
Each SNP was genotyped by using TaqMan SNP Genotyping Assays (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions. The sequences of PCR primers and TaqMan MGB probes used in this study are listed in the Supplementary Table S2. Allele detection and genotype scoring were performed with ABI 7900HT sequence detection system and Sequence Detection System software version 2.2.2 (Applied Biosystems).
Statistical analysis
SNPs were tested individually for the statistical significance of association with drinking behavior by using the _χ_2-test statistic for genotype distributions and allele frequencies. SNPs’ genotype departures from the Hardy–Weinberg equilibrium were tested using a _χ_2-test with 1 degree of freedom. As for SNPs that were shown to be associated with drinking behavior, we performed not only an analysis of variance to evaluate their influences on blood pressure but also an analysis of covariance to control for the confounding factors. In addition, we performed a linear regression analysis that included ALDH2 rs671 genotype, age, BMI, the amounts of drinking and smoking status to determine the predictor of increased blood pressure separately by sex. All statistical analyses were performed by using SPSS 14.0J for Windows.
Results
Characteristics of study subjects and SNPs related to drinking behavior
Characteristics of study subjects are shown in Table 1. The average age of the subjects was 48.8 years and the proportion of male subjects was 60.4%. The percentages of male and female regular drinkers were 55.5 and 14.9%, respectively, and those of male and female nondrinkers were 14.5 and 38.7%, respectively.
Table 1 Clinical characteristics of participants
The results of genotyping are shown in Table 2. We evaluated the influence of 10 tag SNPs (1 from ADH1A, 2 from ADH1B, 3 from ADH1C and 4 from ALDH2) on drinking behavior. In the comparison between regular drinkers and nondrinkers, significant differences were observed for ADH1B Arg47His (rs1229984, G-to-A polymorphism) and for four tag SNPs from ALDH2, including Glu487Lys (rs671, G-to-A polymorphism) in both male and female subjects in all genetic models tested. In addition, modestly significant differences were observed for ADH1A rs2276332 in both sexes and ADH1C rs1693431 in female subjects. Among the four ALDH2 tag SNPs, rs671 showed the most significant association with drinking behavior (_P_=2.9 × 10−251 in male and 3.2 × 10−49 in female subjects by allele frequency comparison) and the other SNPs did not show additional contributions to the association when evaluated by logistic regression (data not shown). It is noted that 22.7% of nondrinkers (both sexes combined) possessed Glu homozygotes (GG genotype) of ALDH2 rs671, whereas no male and female drinkers had Lys homozygotes (AA genotype) of rs671.
Table 2 Test of association between drinking behavior and 10 SNPs selected from four ethanol-metabolizing genes
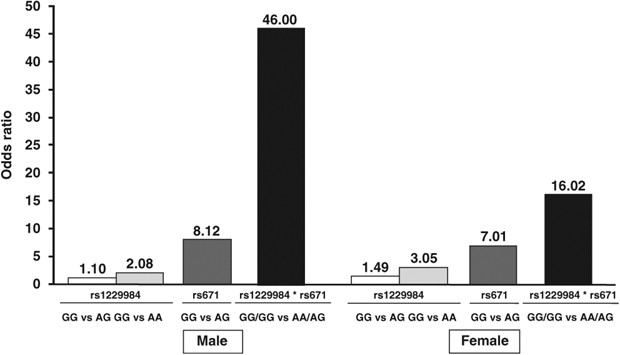
Next, synergistic effects of two significant SNPs on drinking behavior were estimated separately by sex. Subjects with GG genotype of ALDH2 rs671 turned out to be regular drinkers in most (99%) of the cases where they simultaneously possessed AA genotype of ADH1B rs1229984. Thus, the combination of two SNP genotypes appeared to substantially influence drinking behavior (Table 3). When subjects with AA genotype of ADH1B rs1229984 and AG genotype of ALDH2 rs671 were considered as a reference group, the combined odds ratios were calculated to be 46.0 (95% CI, 6.3–334.2) in male and 16.0 (95% CI, 7.63–33.63) in female subjects with double homozygotes of ‘drinking-prone’ allele, that is, G allele in both loci. When odds ratios were compared for the association between each of the two loci or their combination and drinking behavior, the odds ratios for the combination were markedly higher than those for each locus, indicating that the combination of the two SNPs exerts synergistic effect on drinking behavior (Figure 1).
Table 3 Odds ratios for drinking behavior associated with combinations of ADH1B (rs1229984) and ALDH2 (rs671) polymorphisms
Figure 1
Comparison of odds ratios for the association of rs1229984, rs671 and their combination with drinking behavior in male and female subjects. Rs1229984 and rs671 indicate the odds ratios for the respective loci, and rs1229984*rs671 indicates the odds ratio for the combination of the two loci.
Influences of two SNPs related to drinking behavior on blood pressure
The relationship between blood pressure and two SNPs that we found to be significantly associated with drinking behavior was further evaluated in each category of drinking behavior (Table 4). The difference in systolic blood pressure between the subjects with AG genotype of ALDH2 rs671 and those with GG genotype was 1.9 mm Hg (_P_=0.018) and 2.6 mm Hg (_P_=0.0001) for chance drinkers and regular drinkers (⩾1 gou day−1), respectively, in all subjects. The diastolic blood pressure was also significantly higher (1.9 mm Hg, P<0.001) in subjects with GG genotype of ALDH2 rs671 than in those with AG genotype for regular drinkers (⩾1 gou day−1). However, no significant relationship was found between ADH1B rs1229984 and blood pressure in any category.
Table 4 Influence of ADH1B (rs1229984) and ALDH2 (rs671) polymorphisms on blood pressure according to drinking behavior
In the category of male regular drinkers (⩾1 gou day−1), GG genotype of ALDH2 rs671 was found to be an independent predictor of increased systolic (β-coefficient=2.96, _P_=0.03) and diastolic (β-coefficient=2.26, _P_=0.01) blood pressure after adjusting for the amounts consumed at one time (Table 5). Figure 1 shows blood pressure measurements (in y axis) plotted against the amounts of alcohol consumption (in x axis) according to the ALDH2 rs671 genotype (GG vs. AG) to visually show a relationship between the two parameters. In accordance with the results of linear regression analysis (Table 5), there was a rough tendency of blood pressure elevation according to the increased amounts of alcohol consumption in female subjects but not apparently so in male subjects in our population studied.
Table 5 Predictive factors of increasing blood pressure among regular drinkers (⩾1 gou day−1)
Discussion
We have identified through the candidate gene approach that two SNPs, ADH1B rs1229984 and ALDH2 rs671, are commonly found in the Japanese general population and that they significantly influence drinking behavior in a synergistic manner. In addition, it is suggested that ALDH2 rs671 influences the level of blood pressure independently of the amounts of alcohol consumption.
Several studies have been reported to test the genetic association between drinking behavior and candidate gene polymorphisms in the alcohol-metabolizing enzymes such as ADH1B and ALDH2 in alcoholic patients.3, 6 Only a few studies, however, have explored this type of association in a population-based setting. Liu et al.7 performed an association study between drinking behavior and 153 SNPs derived from 40 candidate genes in 339 middle-aged men. In their study, a multivariate analysis showed modest association between drinking behavior and ALDH2 rs671. Owing to the sample limitation (modest size and enrollment of male subjects only), this finding needs to be validated in the large-scale Japanese general population.
It is well known that drinking behavior is principally defined by a combination of factors, that include constitutional intolerance (heritable or genetic factors),3, 6, 7 the status of mental health (psychological factors)8 and the drinking habits of the spouse (environmental factors).9 In this study, after screening of 10 tag SNPs derived from four candidate genes in the alcohol-metabolizing pathway, we have come across two notable SNPs, ADH1B rs1229984 and ALDH2 rs671, that appear to be significantly associated with drinking behavior. In particular, ALDH2 rs671 seems to confer strong constitutional intolerance to alcohol and we have found that there is an inverse relation between the amounts of individual alcohol consumption and the frequency of ‘intolerance-type’ A-allele of rs671 (see Supplementary Figure S2), suggesting the multifactorial regulation of alcohol preference.
The A-allele frequencies of ALDH2 rs671 are 65% (male subjects) and 43% (female subjects) in nondrinkers, and the percentages are relatively concordant among different studies that have enrolled the Japanese general subjects in a population-based setting. It has been reported that the corresponding allele frequencies are 57% (male subjects) and 35% (female subjects) in nondrinkers in two earlier studies,10, 11 where the average age of the participants is 8–11 years older and the population structure (for example, people's lifestyle and profession) is supposed to be different from that in this study. In addition, we have identified the synergistic influences of ALDH2 rs671 and ADH1B rs1229984 to confer alcohol intolerance; the combined odds ratios were calculated to be 46.0 (95% CI, 6.3–334.2) in male subjects and 16.0 (95% CI, 7.63–33.63) in female subjects (Table 3). Matsuo et al.12 also studied the association between drinking behavior and the combination of the SNPs, which were the same as those examined in this study. They found an obvious association between ADH2 minor allele and drinking behavior in the group of subjects heterozygous for ALDH2. The association was stronger than that in the group of subjects having other ALDH2 genotypes. Discrepancies between the results obtained in studies by Matsuo et al.12 and by us may arise from differences in the study population. Matsuo et al.12 conducted the study with outpatients visiting the hospital for the treatment of any disease and not having an earlier history of cancer or alcoholism. These patients might change their drinking behavior on the occasion of their illness. Therefore, it is speculated that the drinking behavior of these patients was different from that of the general population enrolled in our study. A series of earlier studies have reported the functional significance of ALDH2 rs671 and ADH1B rs1229984,13, 14 both of which lead to amino acid substitutions and distinct activities of complementary key enzymes in the alcohol-metabolizing pathway; that is, alcohol dehydrogenase (ADH) that catalyzes the conversion of alcohols to acetaldehydes and aldehyde dehydrogenase (ALDH) that oxidizes acetaldehyde to acetic acid. Thus, a varied combination of factors in individual genetic impacts attributable to ALDH2 rs671 and ADH1B rs1229984 seem to take a substantial part in the regulation of drinking behavior in the population at large, and the genotype information on these two SNPs is useful for predicting individual alcohol preference in the Japanese.
The most interesting finding in our study is that the ALDH2 rs671 genotype can modify the alcohol–blood pressure relationship additionally to alcohol preference. Several studies have investigated the influence of rs671 genotypes on blood pressure.11, 15, 16, 17 Almost all earlier studies, except for the one by Itoh et al., have reported that ALDH2 rs671 cannot significantly influence the alcohol–blood pressure relationship.18 The following reasons may account for discrepancies between the results of our study and those of others: (1) sample size is relatively small in other studies; (2) physiological, aging-related blood pressure elevation is not negligible when the average age of the participants is relatively high in some studies; (3) the confounding pressor effects of the amounts of alcohol consumption may not be sufficiently adjusted for by multivariate analysis alone using the entire study population; and (4) nondrinkers should be excluded from the analysis to evaluate ‘gene–environment (that is, alcohol intake in this case) interaction.’
Blood acetaldehyde concentration is considered to play some role in the involvement of ALDH genotype in the alcohol–blood pressure relationship. It has been known that ALDH2 polymorphism has a strong association with the accumulation of acetaldehyde. When an ethanol loading test was carried out in groups of subjects having three different genotypes of two alleles in the ALDH2 locus, the peak blood concentration of acetaldehyde was higher in the order of the subjects having ALDH2 Lys homozygote, heterozygote and Glu homozygote, and there were significant differences among the three groups.19 In addition, it has been reported that acetaldehyde exerts vasodilating effects, especially with acute administration. A direct vasodilating action of acetaldehyde,20 and indirect vasodilation mediated by the release of prostacyclin from the vascular wall21 and by activation of the kinin system,22 were proposed to be the mechanisms of action of this effect. In addition, concerning the influence of ALDH genotype on the alcohol–blood pressure relationship, Kawano et al.23 showed in their study with hypertensive patients that the acute reduction of blood pressure by ethanol intake was greater in patients who showed facial flush after alcohol intake (ALDH2 Lys homozygotes) than in patients who did not (Glu homozygotes). In this study, we also found a reduction in blood pressure in subjects heterozygous for ALDH2 rs671, suggesting the influence of vasodilation by acetaldehyde. However, it is difficult to conclude that the blood pressure reduction was induced by the acute effect of acetaldehyde because of a lack of information about time after the last drink and the amount of alcohol intake of the subjects in this study. Further study will be required to elucidate the mechanism of the influence of ALDH genotype on the alcohol–blood pressure relationship.
In this study, the influence of ALDH2 rs671 polymorphism on alcohol–blood pressure relationship was found only in men. The reason for the failure to show the elevation of blood pressure in the female counterpart is unclear, but may be due to smaller number of regular drinkers in the female than in the male subject in this study. It is also possible that we were not able to evaluate the influence of the amount of alcohol intake sufficiently because of the lack of detailed information about the amount of individual alcohol intake in terms of ethanol. To verify this result, further studies will be needed with a sufficient number of female subjects and detailed information on their alcohol intake.
There are limitations in this study. First, several epidemiological studies have shown that some dietary factors such as total energy intake, salt consumption and coffee drinking are important determinants of blood pressure,24 but no nutritional information is available in this study. In addition, although we have evaluated the alcohol–blood pressure relationship according to the frequency and the amounts of alcohol intake, it is possible that the drinking pattern (cumulative or periodic) can affect the alcohol–hypertension relationship.25 Moreover, we could not obtain information on the duration of alcohol intake or the time after the last drinking, which may affect the relationship between alcohol consumption and blood pressure. Finally, this study analyzed only casual blood pressure, which may overestimate the pressor affect of alcohol in each individual, especially in the Japanese. These issues warrant further investigation in the context of gene–environment interaction.
In conclusion, our data suggest that not only environmental factors but also genetic factors substantially determine drinking behavior in the Japanese general population. Furthermore, the genetic factors can influence the sensitivity to pressor effects of alcohol. These findings are useful in terms of the individualization of treatment strategies employed to reduce alcohol consumption.
References
- Klatsky AL, Friedman GD, Siegelaub AB, Gerard MJ . Alcohol consumption and blood pressure Kaiser-Permanente Multiphasic Health Examination data. N Engl J Med 1977; 296: 1194–1200.
Article CAS Google Scholar - Muntwyler J, Hennekens CH, Buring JE, Gaziano JM . Mortality and light to moderate alcohol consumption after myocardial infarction. Lancet 1998; 352: 1882–1885.
Article CAS Google Scholar - Goedde HW, Agarwal DP, Fritze G, Meier-Tackmann D, Singh S, Beckmann G, Bhatia K, Chen LZ, Fang B, Lisker R, Paik YK, Rothhammer F, Saha N, Segal B, Srivastava LM, Czeizel A . Distribution of ADH2 and ALDH2 genotypes in different populations. Hum Genet 1992; 88: 344–346.
Article CAS Google Scholar - Thomasson HR, Edenberg HJ, Crabb DW, Mai XL, Jerome RE, Li TK, Wang SP, Lin YT, Lu RB, Yin SJ . Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am J Hum Genet 1991; 48: 677–681.
CAS PubMed PubMed Central Google Scholar - Takeuchi F, Yanai K, Morii T, Ishinaga Y, Taniguchi-Yanai K, Nagano S, Kato N . Linkage disequilibrium grouping of single nucleotide polymorphisms (SNPs) reflecting haplotype phylogeny for efficient selection of tag SNPs. Genetics 2005; 170: 291–304.
Article CAS Google Scholar - Muramatsu T, Wang ZC, Fang YR, Hu KB, Yan H, Yamada K, Higuchi S, Harada S, Kono H . Alcohol and aldehyde dehydrogenase genotypes and drinking behavior of Chinese living in Shanghai. Hum Genet 1995; 96: 151–154.
Article CAS Google Scholar - Liu Y, Yoshimura K, Hanaoka T, Ohnami S, Ohnami S, Kohno T, Yoshida T, Sakamoto H, Sobue T, Tsugane S . Association of habitual smoking and drinking with single nucleotide polymorphism (SNP) in 40 candidate genes: data from random population-based Japanese samples. J Hum Genet 2005; 50: 62–68.
Article CAS Google Scholar - Stranges S, Notaro J, Freudenheim JL, Calogero RM, Muti P, Farinaro E, Russell M, Nochajski TH, Trevisan M . Alcohol drinking pattern and subjective health in a population-based study. Addiction 2006; 101: 1265–1276.
Article Google Scholar - Leonard KE, Mudar P . Peer and partner drinking and the transition to marriage: a longitudinal examination of selection and influence processes. Psychol Addict Behav 2003; 17: 115–125.
Article Google Scholar - Takagi S, Baba S, Iwai N, Fukuda M, Katsuya T, Higaki J, Mannami T, Ogata J, Goto Y, Ogihara T . The aldehyde dehydrogenase 2 gene is a risk factor for hypertension in Japanese but does not alter the sensitivity to pressor effects of alcohol: the Suita study. Hypertens Res 2001; 24: 365–370.
Article CAS Google Scholar - Amamoto K, Okamura T, Tamaki S, Kita Y, Tsujita Y, Kadowaki T, Nakamura Y, Ueshima H . Epidemiologic study of the association of low-Km mitochondrial acetaldehyde dehydrogenase genotypes with blood pressure level and the prevalence of hypertension in a general population. Hypertens Res 2002; 25: 857–864.
Article CAS Google Scholar - Matsuo K, Wakai K, Hirose K, Ito H, Saito T, Tajima K . Alcohol dehydrogenase 2 His47Arg polymorphism influences drinking habit independently of aldehyde dehydrogenase 2 Glu487Lys polymorphism: analysis of 2299 Japanese subjects. Cancer Epidemiol Biomarkers Prev 2006; 15: 1009–1013.
Article CAS Google Scholar - Matsuo Y, Yokoyama R, Yokoyama S . The genes for human alcohol dehydrogenases beta 1 and beta 2 differ by only one nucleotide. Eur J Biochem 1989; 183: 317–320.
Article CAS Google Scholar - Yoshida A, Huang IY, Ikawa M . Molecular abnormality of an inactive aldehyde dehydrogenase variant commonly found in Orientals. Proc Natl Acad Sci USA 1984; 81: 258–261.
Article CAS Google Scholar - Saito K, Yokoyama T, Yoshiike N, Date C, Yamamoto A, Muramatsu M, Tanaka H . Do the ethanol metabolizing enzymes modify the relationship between alcohol consumption and blood pressure? J Hypertens 2003; 21: 1097–1105.
Article CAS Google Scholar - Hashimoto Y, Nakayama T, Futamura A, Omura M, Nakarai H, Nakahara K . Relationship between genetic polymorphisms of alcohol-metabolizing enzymes and changes in risk factors for coronary heart disease associated with alcohol consumption. Clin Chem 2002; 48: 1043–1048.
CAS PubMed Google Scholar - Yamada Y, Sun F, Tsuritani I, Honda R . Genetic differences in ethanol metabolizing enzymes and blood pressure in Japanese alcohol consumers. J Hum Hypertens 2002; 16: 479–486.
Article CAS Google Scholar - Itoh T, Matsumoto M, Nakamura M, Okada A, Shirahashi N, Hougaku H, Hashimoto H, Sakaguchi M, Handa N, Takeshita T, Morimoto K, Hori M . Effects of daily alcohol intake on the blood pressure differ depending on an individual's sensitivity to alcohol: oriental flushing as a sign to stop drinking for health reasons. J Hypertens 1997; 15: 1211–1217.
Article CAS Google Scholar - Harada S . Dispersal of the ALDH2 mutant in Mongoloid population. In: Akazawa T, Szathmary EJE (eds). Prehistoric Dispersal of Mongoloid. Oxford University Press: London, 1995, pp 165–171.
Google Scholar - Altura BM, Altura BT . Microvascular and vascular smooth muscle actions of ethanol, acetaldehyde, and acetate. Fed Proc 1982; 41: 2447–2451.
CAS PubMed Google Scholar - Guivernau M, Baraona E, Lieber CS . Acute and chronic effects of ethanol and its metabolites on vascular production of prostacyclin in rats. J Pharmacol Exp Ther 1987; 240: 59–64.
CAS PubMed Google Scholar - Hatake K, Taniguchi T, Ouchi H, Sakaki N, Hishida S, Ijiri I . Possible involvement of kinins in cardiovascular changes after alcohol intake. Pharmacol Biochem Behav 1990; 35: 437–442.
Article CAS Google Scholar - Kawano Y, Abe H, Kojima S, Ashida T, Yoshida K, Imanishi M, Yoshimi H, Kimura G, Kuramochi M, Omae T . Acute depressor effect of alcohol in patients with essential hypertension. Hypertension 1992; 20: 219–226.
Article CAS Google Scholar - Beilin LJ, Puddey IB, Burke V . Lifestyle and hypertension. Am J Hypertens 1999; 12: 934–945.
Article CAS Google Scholar - Rakic V, Puddey IB, Burke V, Dimmitt SB, Beilin LJ . Influence of pattern of alcohol intake on blood pressure in regular drinkers: a controlled trial. J Hypertens 1998; 16: 165–174.
Article CAS Google Scholar
Acknowledgements
This work was supported by a grant for Core Research for the Evolutional Science and Technology from the Japan Science Technology Agency. N Kato is a principal investigator and was responsible for the whole study including the design, experimentation and data interpretation.
Author information
Authors and Affiliations
- Division of Genomic Epidemiology, Department of Clinical Research and Informatics, Research Institute, International Medical Center of Japan, Tokyo, Japan
Miyuki Tsuchihashi-Makaya - Department of Gene Diagnostics and Therapeutics, Research Institute, International Medical Center of Japan, Tokyo, Japan
Masakuni Serizawa, Kazuyuki Yanai & Norihiro Kato - Department of Geriatric Medicine, Osaka University Graduate School of Medicine, Osaka, Japan
Tomohiro Katsuya & Toshio Ogihara - Wellcome Trust Sanger Institute, Cambridge, UK
Fumihiko Takeuchi - Amagasaki Health Medical Foundation, Amagasaki, Japan
Akihiro Fujioka - Collaborating Center for Prevention of Cardiovascular Diseases, Kyoto, Japan
Yukio Yamori
Authors
- Miyuki Tsuchihashi-Makaya
You can also search for this author inPubMed Google Scholar - Masakuni Serizawa
You can also search for this author inPubMed Google Scholar - Kazuyuki Yanai
You can also search for this author inPubMed Google Scholar - Tomohiro Katsuya
You can also search for this author inPubMed Google Scholar - Fumihiko Takeuchi
You can also search for this author inPubMed Google Scholar - Akihiro Fujioka
You can also search for this author inPubMed Google Scholar - Yukio Yamori
You can also search for this author inPubMed Google Scholar - Toshio Ogihara
You can also search for this author inPubMed Google Scholar - Norihiro Kato
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toMiyuki Tsuchihashi-Makaya.
Additional information
Supplementary Information accompanies the paper on Hypertension Research website (http://www.nature.com/hr)
Supplementary information
Rights and permissions
About this article
Cite this article
Tsuchihashi-Makaya, M., Serizawa, M., Yanai, K. et al. Gene–environmental interaction regarding alcohol-metabolizing enzymes in the Japanese general population.Hypertens Res 32, 207–213 (2009). https://doi.org/10.1038/hr.2009.3
- Received: 07 July 2008
- Revised: 01 October 2008
- Accepted: 04 November 2008
- Published: 20 February 2009
- Issue Date: March 2009
- DOI: https://doi.org/10.1038/hr.2009.3