Inhibition of biofilm formation in Bacillus subtilis by new halogenated furanones (original) (raw)
Introduction
Among the causes of the infections, including hospital-acquired infections, Gram-positive bacteria are important contributors. Their resistance both to antibiotics and to the human immune system that significantly limits the therapeutic effects of antibiotics is often associated with their location in biofilm.1,2 Bacilllus biofilms are important contaminants in food industry plants, and they are also being widely used as a model system for biofilm-related studies.3,4
In the last decade, the natural and synthetic furanones were noted as biofilm-inhibiting agents.5,6 First, the natural furanone from the red algae Delisea pulchra, (5_Z_)-4-bromo-5-(bromomethylene)-3-butyl-2(5_H_)-furanone, was shown to inhibit the growth, swarming and biofilm formation of Bacillus subtilis.5,7,8 The influence of furanones on opportunistic and pathogenic bacteria and yeast was also investigated.9,10 It is known that furanones inhibit the quorum sensing of Gram-negative bacteria by affecting both the species-specific11,12 and the non-species-specific12 autoinducer signals AI-1 (acylated homoserine lactones) and AI-2, respectively.13,14 However, very little is known about the mechanisms of the growth and of the biofilm formation inhibition by furanones in Gram-positive bacteria.
In B. subtilis, several transcription factors control the quorum-determined responses. The transcription factor SinR is the master regulator redirecting B. subtilis from the planktonic form to biofilm formation.15,16 In the exponentially growing cells, SinR constitutively represses the biofilm-related genes.17 When nutrient limitation is sensed, SinI activity increases and antagonizes the SinR. This results in the loss of motility and in biofilm production. Before the discovery of SinR as the master regulator of biofilm formation in B. subtilis, Spo0A and σH were identified as transcriptional regulators required for biofilm development.15,18,19 The yveK-T yvfA-F operon, later renamed epsA-O, was predicted to encode products likely to be involved in synthesis and export of the extracellular polymeric substance, or EPS (exopolysaccharide), being under control of both Spo0A and σH.15 It was also shown that SinR binding to the eps regulatory region is inhibited by complex formation with SinI in vitro.16 Thus, SinR acts as a transcriptional repressor of the genes involved in producing the extracellular matrix, and SinI can antagonize its action.
Here we introduce the new derivatives of 2(5_H_)-furanones that efficiently inhibit the growth and the biofilm formation by B. subtilis, apparently via the repression of the biofilm-related regulatory pathways and probably affecting Spo0A transcriptional regulator.
Experimental procedure
Furanones
The structures of all studied 2(5_H_)-furanone derivatives are presented in Figure 1. Mucochloric acid (3,4-dichloro-5-hydroxy-2(5_H_)-furanone) F1 (Shostka Chemical Reagents Plant, Shostka, Ukraine) was commercially available and was further recrystallized from water, m.p. 127 °C. Mucobromic acid (3,4-dibromo-5-hydroxy-2(5_H_)-furanone) F2 (Sigma Aldrich, USA) was used without further purification. 3,4-Dichloro-5-[(1,3-dichloropropan-2-yloxy)]-2(5_H_)-furanone F8,20 3-chloro-5-hydroxy-4-[(4-methylphenylsulfanyl)]-2(5_H_)-furanone F12,21 3-chloro-5-hydroxy-4-[(4-methylphenylsulfonyl)]-2(5_H_)-furanone F7022 and 3,4-dichloro-5-[(4-methylphenylsulfonyl)]-2(5_H_)-furanone F7622 were synthesized according to the previously known procedures. 3-Chloro-5-[(2-chloroethoxy)]-4-[(4-methylphenylsulfanyl)]-2(5_H_)-furanone F21 (Hoang Thi Lien, 2015, unpublished data) and 3-bromo-5-methoxy-4-[(4-methylphenylsulfanyl)]-2(5_H_)-furanone F23 (Kosolapova, 2015, unpublished data) have not been described earlier. The furanones were dissolved and diluted in dimethyl sulfoxide to a final stock concentration of 20 mg ml−1 and stored at −20 °C.
Figure 1
Chemical structures of furanones used in this study.
Bacterial strains, plasmids and growth conditions
B. subtilis 168 (wild type) and its mutant strains shown below were used. B. subtilis K444 (trpC2 amyE::PyqxM-lacZ (cat)), B. subtilis K511 (trpC2 amyE::Peps-gfp (cat)), B. subtilis WTF92 (sinR::cat), B. subtilis W1274 (sinI::spc), B. subtilis WTF96 (spo0A::cat) and B. subtilis W1050 (abrB::kan) were obtained from Professor Dr Kazuo Kobayashi.23 B. subtilis JMS682 (amyE::srfA-lacZΩ682 (neo)) was provided by Professor Dr Alan Grossman.24 Escherichia coli BL21 (DE3) was used for protein overexpression. Plasmids pET15b-TnrA, pGP176 and pDG-GlnK-ST for the overproduction of TnrA-His6, GlnK-ST and glutamine synthetase (GS), respectively, had been constructed previously.25 B. subtilis GP250 (trpC2 amyE::Pnrg-lacZ (kan)) was obtained from Professor Dr J. Stulke.26 B. subtilis and E. coli strains were maintained and grown on the Luria-Bertani (LB) medium. The biofilm formation was examined on the modified Basal medium (BM) containing glucose 5 g, peptone 7 g, MgSO4 7 H2O 2.0 g and CaCl2 0.05 g in 1.0 l tap water. Spizizen minimal medium (SMM) was prepared as described previously27 and supplemented with 20 mM potassium nitrate as a sole nitrogen source to provide the nitrogen-limited conditions.27 Antibiotics were used at following concentrations: ampicillin, 100 μg ml−1; chloramphenicol, 10 μg ml−1; kanamycin, 10 μg ml−1; and spectinomycin, 60 μg ml−1.
Determination of the MIC
The MICs of furanones against B. subtilis were determined by the microdilution method in BM. Furanones were diluted by BM in a 96-well microtiter plate to final concentrations ranging from 1 to 100 μg ml−1 (1.0, 2.5, 5.0, 10.0, 25.0, 50.0, 100.0 μg ml−1). A 250-μl aliquot of the bacterial suspension (2–9 × 106 CFU ml−1) was inoculated and incubated at 37 °C over 24 h. The MIC was determined as the lowest concentration that completely inhibited bacterial growth. The minimal biofilm-inhibiting concentration was defined as the concentration that significantly repressed biofilm formation (OD570<0.1).
Crystal violet assay
The biofilm formation was studied using crystal violet staining assay as previously described.28 Briefly, a 300-μl aliquot of the B. subtilis cells' suspension (2–9 × 106 CFU ml−1) was inoculated in BM and grown during 72 h in 96-well microtiter plate. Then, the culture supernatant was discarded, and the wells were washed twice with phosphate-buffered saline to remove nonadherent cells. The plates were air dried for 20 min, and the surface-attached cells were stained with 300 μl of 0.1% crystal violet for 20 min. Subsequently, the crystal violet was removed and the plate was washed three times with pure water. After 30 min of air drying, 300 μl of 96% ethanol was added to resolubilize bound crystal violet, and the absorbance was read at 570 nm using the microplate reader Tecan Infinite 200 Pro (Tecan Austria GmbH, Austria). Wells incubated with the cell-free medium were also stained and served as a blank.
β-Galactosidase and glutamine synthetase (GS) assays
The β-galactosidase activity was measured by the Miller’s protocol29 with modifications.30 Cells were collected, resuspended in 400 μl of Z-buffer (60 mM Na2HPO4 • 7 H2O, 40 mM NaH2PO4 • H2O, 10 mM KCl and 1 mM MgSO4 • 7 H2O (pH 7.0)) containing 2 mg ml−1 lysozyme and incubated for 10 min at 37 °C. Then, 400 μl of Z-M buffer containing additionally 0.005% cetyl trimethylammonium bromide (CTAB) and 50 mM β-mercaptoethanol were added. After preincubation at 30 °C for 5 min, the reaction was started by adding of 160 μl of 4 mg ml−1 o-nitrophenyl-β-D-galactopyranoside in Z-buffer. When the yellow color appeared, the reaction was stopped by the addition of 400 μl of 1 M Na2CO3. For the blank solution, the Na2CO3 was added before the incubation. The β-galactosidase activity was measured at A420 nm. To calculate the Miller units, we used the following formula: [A420/(A600 of 1:10 dilution of cells × time of incubation)] × 1000.
GS activity was measured by the biosynthetic assay.30 Cells were collected, resuspended in 450 μl of the reaction mixture consisted of 100 mM imidazole-HCl (pH 7.0), 125 mM hydroxylamine-HCl, 20 mM MgCl2·6 H2O, 10 mM ATP, 0.005% CTAB and 0.1 mg ml−1 lysozyme and were pre-incubated at 37 °C for 10 min to permeabilize the cells. The reaction was initiated by adding 50 μl of 500 mM glutamate, giving a final assay volume of 500 μl. After a 20-min incubation, the reaction was stopped by the additon of 1 ml of stop mix (5.5% FeCl3 • 6 H2O (w/v), 2% trichloroacetic acid (w/v) and 0.78% HCl) and centrifugation for 5 min at 10 000 g to remove the precipitate. The Fe (III) complex of the reaction product γ-glutamylhydroxamate was measured spectrophotometrically by absorbance at 540 nm. GS-specific activity was calculated as nmol of γ-glutamylhydroxamate produced per minute A600 per cell. A reaction mixture without glutamate served as the blank.
Proteins' preparation and manipulation
TnrA from B. subtilis 168 cells carrying His6-tag on its N-terminus was overproduced in E. coli BL21 strain using pET15b expression vector (Novagene, USA) and purified on Ni-NTA column to an apparent electrophoretic homogeneity as described previously.25 Strep-tagged GlnK and GS were overproduced in E. coli BL21 strain using pDG148 expression vector and purified using Strep-tactin column (IBA, Göttingen, Germany) as described previously.25 The influence of furanones on the TnrA interaction with either GlnK or GS was investigated by pull-down analysis as described previously.30,31
Statistics
Experiments were carried out in biological triplicates (that is, newly prepared cultures and medium) with three independent repeats in each one. Biomasses from crystal violet staining as well as enzymes' activities in different experiments were compared using Wilcoxon signed-rank test (paired difference test). Differences were considered significant at P<0.05.
Results
The repression of B. subtilis growth and biofilm formation
It has been shown earlier that adding (5_Z_)-4-bromo-5-(bromomethylene)-3-butyl-2(5_H_)-furanone from the red marine algae D. pulchra results in the twofold inhibition of the B. subtilis cells' growth at concentration 5 μg ml−1 without killing the cells.7 Six novel synthesized furanones (Figure 1) were tested for their ability to repress the growth and the biofilm formation by B. subtilis cells. Mucochloric acid F1 and mucobromic acid F2 were the starting materials for the preparation of all other compounds and thus were used as references. The influence of furanones on the B. subtilis growth as well as on the biofilm formation is shown in Table 1. Although the minimal biofilm-inhibiting concentration for mucochloric acid (F1) was established to be 25 μg ml−1, F2, F8 and F12 completely suppressed the biofilm formation at 10 μg ml−1. Other tested compounds did not exhibit any significant antibacterial activity at low concentrations and therefore were out of further investigation.
Table 1 Concentrations of furanones required for complete inhibition of growth (MIC) and for the biofilm formation (MBIC) by B. subtilis 168
To evaluate the biofilm formation inhibition by furanones, we measured the activity of β-galactosidase expressed from the yqxM promoter, which is activated when cells switch to the biofilm formation.23 B. subtilis K444 cells were grown 72 h in BM without shaking in the presence of F1, F2, F8 and F12 at their determined minimal biofilm-inhibiting concentrations given above. In the presence of all furanones, the activity of β-galactosidase was significantly reduced confirming the repression of transcription from the yqxM promoter (Table 1). In addition, the green fluorescent protein gene expression from the eps promoter in B. subtilis K511 strain was studied by fluorescent microscopy on a Karl Zeiss Axio Imager (Zeiss, Optek, Germany) 2.0 microscope (Supplementary Figure S1). In the presence of F1 (25 μg ml−1), F2, F8 and F12 (10 μg ml−1), a strong decrease in the green fluorescent protein fluorescence was observed that confirms the inhibition of the eps operon expression.
Finding potential cellular targets of furanones
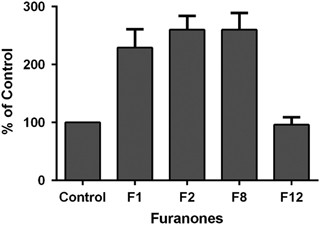
The cellular targets of furanones in Gram-positive bacteria still remain unknown. In some of the previous investigations, the ClpP protease, transcription regulator ComA and some other regulatory proteins were proposed as potential targets for furanones.8 ComA is a transcription factor regulating the competence quorum processes in B. subtilis.32,33 We measured the activity of β-galactosidase expressed from the ComA-dependent srf promoter in B. subtilis JMS682 strain under conditions required for genetic competence development.34 In the presence of F1, F2 and F8 at their minimal biofilm-inhibiting concentrations, the activity of β-galactosidase was twofold higher compared with the control indicating the activation of ComP–ComA regulatory pathway; F12 had almost no impact on the β-galactosidase activity (Figure 2).
Figure 2
Influence of furanones on the activity of β-galactosidase expressed from the ComA-dependent srfA promoter in B. subtilis JMS682.
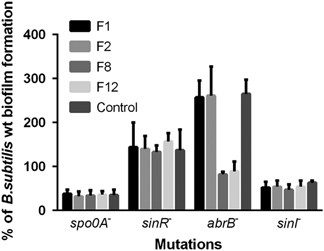
In further attempts to identify the intracellular targets of furanones, we tested the biofilm formation in the presence of furanones by B. subtilis strains lacking Spo0A, AbrB, SinI and SinR transcriptional factors involved in the biofilm formation.15,16 Furanones F2, F8 and F12 were added up to the final concentration of 5 μg ml−1 and F1 up to 15 μg ml−1, which led to approximately double inhibition of the biofilm formation. The residual biofilm formation in the absence and in the presence of furanones is shown in Figure 3.
Figure 3
Biofilm formation in the presence of furanones by strains lacking transcription factors involved in biofilm formation control (Spo0A, SinR, AbrB and SinI). The biofilm formation by B. subtilis strains in the absence of furanones served as a control and was defined as 100%.
The biofilm formation by the Spo0A- and SinI-deficient strains was low (almost at the borderline of the method sensitivity) and no repression by any furanones could be observed. The SinR-deficient strain exhibited twofold increase in the biofilm formation independently of the presence of furanones, indicating that these compounds did not affect the SinR–SinI regulatory pathway. Unexpectedly, in the absence of the AbrB, F8 and slightly F12 decreased the biofilm formation.
The effect of furanones on the nitrogen metabolism of B. subtilis
In the course of our investigations, we found that the growth of B. subtilis 168 in minimal medium (SMM) was considerably slower in the presence of F12 under conditions of nitrogen starvation (not shown). In exponentially growing nitrogen-limited cells, we found 10-fold lower activities of GS and β-galactosidase expressed from the nrgA promoter, which is activated under nitrogen-limited conditions26 (Table 2, in situ measurements). As a control, the influence of furanones on activities of GS and β-galactosidase was investigated in vitro. For that, the extract of the cells grown without any furanone was pre-incubated with compounds and the activities were measured (Table 2). Only F12 slightly affected β-galactosidase and GS in vitro, indicating that the observed repression of their activities is indeed an effect of furanones on the cellular metabolism.
Table 2 Influence of furanones on the activities of GS and β-galactosidase expressed from the TnrA-dependent nrgA promoter in B. subtilis GP250
In B. subtilis, the activity of the transcription factor TnrA, the master regulator of nitrogen metabolism, that is bound to the PII-like protein GlnK at the active form,25,26,31 is repressed by the interaction with the feedback-inhibited GS. In the absence of its partner proteins, TnrA is eliminated by proteolysis.27 To investigate the possible influence of furanones on the nitrogen metabolism, we checked TnrA interaction with both GlnK and GS by pull-down analysis in the presence of furanones, as described in Supplementary Material. F1, F2 and F8 did not affect proteins' interactions in vitro and TnrA was co-eluted with both GlnK and GS despite the presence of furanones (Figures 4a and b). In contrast, the furanone F12 impaired the interaction of TnrA with either GlnK or GS in a dose-dependent manner (Figures 4a and b). F12 (10 and 20 μg ml−1) abrogated completely TnrA binding to the GS and GlnK, respectively.
Figure 4
The influence of furanones on the interaction of transcription factor TnrA with the PII-like protein GlnK (a) and the GS (b). TnrA was premixed with the strep-tagged GlnK or GS in the presence of furanone, loaded onto the strep-tactin sepharose and eluted. The elution fractions were further analyzed by using anti-TnrA antibodies.
Discussion
B. subtilis, like many other bacteria, forms biofilms that make them resistant to biocides and to the immune system of the host. In nature, the marine algae produces furanones to prevent its colonization by bacteria.12 These natural furanones and their derivatives completely repressed the biofilm formation at the concentrations of about 40–50 μg ml−1. Here we show that the new derivatives of furanones—F8 and F12—effectively block this process at a concentration of 10 μg ml−1. Therefore, they could be potentially used to prevent the biofilm formation by Bacilli either on various surfaces or in mixed biofilms as shown recently.35
It has been shown previously that furanones interfere with quorum sensing processes of bacteria being very similar to the homoserine lactone, the signal molecule of the AI-I system.11 B. subtilis does not have such a system and cellular targets of furanones remain unidentified. The transcription factor ComA, that is responsible for the quorum-dependent processes in B. subtilis,33 was activated by furanones (Figure 2). Probably, this is related to the stress response adaptation reminiscent to the extracellular enzymes' overproduction controlled by DegU-DegS two-component signal transduction system.36,37 These data indicate that furanones do not affect the competence quorum processes in B. subtilis and, probably, could even increase the lateral gene transfer. Apparently, Firmicutes, at least B. subtilis, are affected by furanones in another way. Our results suggest that the regulatory pathways linked to the Spo0A global regulator could be the putative target (Figure 3). In the absence of the AbrB-dependent repression, the biofilm formation increases, but is repressed in the presence of F8 and F12, whereas AbrB is believed to repress the biofilm-related genes as well as the SinR transcriptional regulator.15,17 This fact could be explained either by the SinR repression that is not being released or by the repression of the production/transduction of the biofilm formation signal. As no effect of the furanones was found in the strains lacking SinI or SinR, F8 and F12 could repress the initiation of biofilm formation. We suggest the Spo0A regulatory pathway as a potential target of these furanones. The absence of their effect in the Spo0A mutant could be the consequence of either strong repression of biofilm-related genes by AbrB in the absence of its cognate antagonist Spo0A or the effect of the low sensitivity of the method (Figure 3).
We also found that the growth of B. subtilis 168 is repressed by F12 under nitrogen starvation conditions. Therefore, it was hypothesized that this compound could also repress the nitrogen metabolism of cells. In the course of our investigations, we found that F12 suppresses the interaction between the master regulator of nitrogen metabolism TnrA in B. subtilis with GS and GlnK protein in vitro. Therefore, likely retards the growth in the nitrogen-limited medium (Figure 4). Probably, in the presence of F12, TnrA does not interact tightly with those interaction partners in vivo, which affect its activity and the whole nitrogen metabolism.30,31 Other furanones did not demonstrate clear dose-dependent effect and therefore did not influence significantly that interaction indicating that this effect is specific for F12. Notably, F12 also suppressed the growth on the nitrogen-rich medium like BM when TnrA and GS are not initially active in the cell. Perhaps, furanone F12 is not specific for the TnrA or GS, and can interact with many other proteins by hydrophobic _p_-tolyl group (see structure on Figure 1). Therefore, the suppression of the regulatory proteins' interaction could be a potential mechanism of the inhibition of the biofilm formation and bacterial growth by furanones. In addition to abolishing the TnrA–GS and TnrA–GlnK interactions, F12 also likely affects the Spo0A interaction with partner proteins that results in the constitutive repression of the biofilm-related genes.
The targets of mucochloric acid F1 and of mucobromic acid F2 still remain unknown. As these furanones demonstrated rather unspecific activity, we cannot exclude that they do not have a localized target but rather affect the whole bacterial metabolism.
References
- Davies D. G . et al. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280, 295–298 (1998).
Article CAS Google Scholar - Mah T. F . O’Toole G. A . Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9, 34–39 (2001).
Article CAS Google Scholar - Vlamakis H . Chai Y . Beauregard P . Losick R . Kolter R . Sticking together: building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 11, 157–168 (2013).
Article CAS Google Scholar - Frank J. F . Walker J . Surman. S . Jass J . (eds) in Industrial Biofouling 205–224 (John Wiley & Sons Ltd, Chicester, NY, USA, 2000).
Google Scholar - Kjelleberg S . Steinberg P . Surface warfare in the sea. Microbiol. Today 28, 134–145 (2001).
Google Scholar - Lonn-Stensrud J . Landin M. A . Benneche T . Petersen F. C . Scheiel A. A . Furanones, potential agents for preventing Staphylococcus epidermidis biofilm infections. J. Antimicrob. Chemother. 63, 309–316 (2009).
Article CAS Google Scholar - Ren D . Sims J. J . Wood T. K . Inhibition of biofilm formation and swarming of Bacillus subtilis by (5Z)-4-bromo-5-(bromomethylene)- 3-butyl-2(5H)-furanone. Lett. Appl. Microbiol. 34, 293–299 (2002).
Article CAS Google Scholar - Ren D . et al. Differential gene expression to investigate the effect of (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone on Bacillus subtilis. Appl. Environ. Microbiol. 70, 4941–4949 (2004).
Article CAS Google Scholar - Margulis A. B . Kolpakov A. I . Ilinskaya O. N . Comparative effect of homoserine lactones and hexylresorcinol in the induction of phenotypic dissociation of bacteria. Microbiology 75, 401–404 (2006).
Article CAS Google Scholar - Duo M . Zhang M . Luk Y. Y . Ren D . Inhibition of Candida albicans growth by brominated furanones. Appl. Microbiol. Biotechnol. 85, 1551–1563 (2010).
Article CAS Google Scholar - Manefield M . et al. Evidence that halogenated furanones from Delisea pulchra inhibit acylatedhomoserine lactone (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein. Microbiology 145, 283–291 (1999).
Article CAS Google Scholar - Ren D . Sims J. J . Wood T. K . Inhibition of biofilm formation and swarming of Escherichia coli by (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone. Environ. Microbiol. 3, 731–736 (2001).
Article CAS Google Scholar - Fuqua W. C . Winans S. C . Greenberg E. P . Quorum sensing in bacteria: the LuxR–LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176, 269–275 (1994).
Article CAS Google Scholar - Vendeville A . Winzer K . Heurlier K . Tang C. M . Hardie K. R . Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 3, 383–396 (2005).
Article CAS Google Scholar - Branda S. S . Gonzalez-Pastor J. E . Ben-Yehuda S . Losick R . Kolter R . Fruiting body formation by Bacillus subtilis. Proc. Natl Acad. Sci. USA 98, 11621–11626 (2001).
Article CAS Google Scholar - Kearns D. B . Losick R . Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev. 19, 3083–3094 (2005).
Article CAS Google Scholar - Branda S. S . Chu F . Kearns D. B . Losick R . Kolter R . A major protein component of the Bacillus subtilis biofilm matrix. Mol. Microbiol. 59, 1229–1238 (2005).
Article Google Scholar - Hamon M. A . Lazazzera B. A . The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol. Microbiol. 42, 1199–1209 (2001).
Article CAS Google Scholar - Britton R. A . et al. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J. Bacteriol. 184, 4881–4890 (2002).
Article CAS Google Scholar - Rapos P . Winternitz P . Beska E . Sohler E . Pesticidal 3,4-dichloro-2(5H)-furanones. CS 138821 19701115 (Czech.) (1970). Chem. Abstr. 76, 34093 (1972).
Google Scholar - Kurbangalieva A. R . et al. Synthesis of novel arylthio derivatives of mucochloric acid. Phosphorus Sulfur Silicon Relat. Elem. 182, 607–630 (2007).
Article CAS Google Scholar - Latypova L. Z . et al. Thioethers, sulphones and sulphoxides of 2(5_H_)-furanone series: synthesis and structure. Russ. J. Org. Chem. 50, 521–534 (2014).
Article CAS Google Scholar - Kobayashi K . SlrR/SlrA controls the initiation of biofilm formation in Bacillus subtilis. Mol. Microbiol. 69, 1399–1410 (2008).
Article CAS Google Scholar - Auchtung J. M . Lee C. A . Grossman A. D . Modulation of the ComA-dependent quorum response in Bacillus subtilis by multiple Rap proteins and Phr peptides. J. Bacteriol. 188, 5273–5285 (2006).
Article CAS Google Scholar - Heinrich A . et al. Interaction of the membrane-bound GlnK-AmtB complex with the master regulator of nitrogen metabolism TnrA in Bacillus subtilis. J. Biol. Chem. 281, 34909–34917 (2006).
Article CAS Google Scholar - Detsch C . Stulke J . Ammonium utilization in Bacillus subtilis: transport and regulatory functions of NrgA and NrgB. Microbiology 149, 3289–3297 (2003).
Article CAS Google Scholar - Kayumov A . Heinrich A . Sharipova M . Iljinskaya O . Forchhammer K . Inactivation of the general transcription factor TnrA in Bacillus subtilis by proteolisis. Microbiology 154, 2348–2355 (2008).
Article CAS Google Scholar - Croes S . et al. Staphylococcus aureus biofilm formation at the hysiologic glucose concentration depends on the _S. aureus_lineage. BMC Microbiol. 9, 229 (2009).
Article Google Scholar - Miller J . Experiments in Molecular Genetics 352–355 (Cold Spring Harbor Laboratory, NY, USA, 1972).
Google Scholar - Fedorova K. P . Kayumov A. R . Woyda K, Ilinskaja O. N . Forchhammer K . Transcription factor TnrA inhibits the biosynthetic activity of glutamine synthetase in Bacillus subtilis. FEBS Lett. 587, 1293–1298 (2013).
Article CAS Google Scholar - Kayumov A . Heinrich A . Fedorova K . Ilinskaya O . Forchhammer K . Interaction of the general transcription factor TnrA with the PII-like protein GlnK and glutamine synthetase in Bacillus subtilis. FEBS J. 278, 1779–1789 (2011).
Article CAS Google Scholar - Msadek T . Kunst F . Rapoport G . in Bacillus subtilis and other Gram-positive Bacteria: Biochemistry, Physiology and Molecular Genetics (eds Sonenshein, A. L., Hoch, J. A., Losick, R.) 713–726 (American Society for Microbiology: Washington DC, USA, 1993).
- Lopez D . Vlamakis H . Kolter R . Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol. Rev. 33, 152–163 (2009).
Article CAS Google Scholar - Anagnostopolous C . Spizizen J . Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81, 741–746 (1961).
Google Scholar - Bridier A . et al. Biofilms of a Bacillus subtilis hospital isolate protect Staphylococcus aureus from biocide action. PLoS ONE 7, e44506 (2012).
Article CAS Google Scholar - Kunst F . Rapoport G . Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 175, 2403 —2407 (1995).
Article Google Scholar - Kayumov A. R . Balaban N. P . Mardanova A. M . Kostrov S. V . Sharipova M. R . Biosynthesis of the subtilisin-like serine proteinase of Bacillus intermedius under salt stress conditions. Microbiology 75, 557–562 (2006).
Article CAS Google Scholar
Acknowledgements
We would like to acknowledge the financial support of this work by the Russian Government Program of Competitive Development of Kazan Federal University, by the Ministry of the Education and Science of the Russian Federation (basic state assignment No 2014/187) and by the RFBR (project No 14-04-31635).
Author information
Authors and Affiliations
- Department of Genetics, Kazan (Volga Region) Federal University, Kazan, Russia
Airat R Kayumov, Elvina N Khakimullina, Irshad S Sharafutdinov, Elena Y Trizna & Anna B Margulis - A.M. Butlerov Institute of Chemistry, Kazan (Volga Region) Federal University, Kazan, Russia
Lilia Z Latypova, Hoang Thi Lien & Almira R Kurbangalieva - Biomedical Engineering Research Center, St Petersburg State Electrotechnical University, St Petersburg, Russia
Mikhail I Bogachev
Authors
- Airat R Kayumov
You can also search for this author inPubMed Google Scholar - Elvina N Khakimullina
You can also search for this author inPubMed Google Scholar - Irshad S Sharafutdinov
You can also search for this author inPubMed Google Scholar - Elena Y Trizna
You can also search for this author inPubMed Google Scholar - Lilia Z Latypova
You can also search for this author inPubMed Google Scholar - Hoang Thi Lien
You can also search for this author inPubMed Google Scholar - Anna B Margulis
You can also search for this author inPubMed Google Scholar - Mikhail I Bogachev
You can also search for this author inPubMed Google Scholar - Almira R Kurbangalieva
You can also search for this author inPubMed Google Scholar
Corresponding authors
Correspondence toAirat R Kayumov or Almira R Kurbangalieva.
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Kayumov, A., Khakimullina, E., Sharafutdinov, I. et al. Inhibition of biofilm formation in Bacillus subtilis by new halogenated furanones.J Antibiot 68, 297–301 (2015). https://doi.org/10.1038/ja.2014.143
- Received: 14 April 2014
- Revised: 12 August 2014
- Accepted: 16 September 2014
- Published: 22 October 2014
- Issue Date: May 2015
- DOI: https://doi.org/10.1038/ja.2014.143