Imaging Drosophila gene activation and polymerase pausing in vivo (original) (raw)
Main
Determining the complete genome sequence of humans, Drosophila and other model higher eukaryotes was an important step in cataloguing the complete code that programmes the development and maintenance of multicellular organisms. A critical challenge that remains is to determine in molecular terms how this code is read and regulated in higher eukaryotes1. The regulation of transcription of the genome is a major mode by which an organism controls both its homeostasis and development. This regulation is executed mainly through the interactions of a plethora of transcription factor proteins and RNAs with each other and with DNA and associated histones2. These interactions then dictate when, where, and to what level specific genes are transcribed. Although biochemical and genetic approaches have identified many of the macromolecular players and their activities, a mechanistic understanding of how they operate in gene regulation can be guided and tested by direct imaging of these molecular interactions and the resulting biochemical processes in living cells.
Two developments in the imaging of transcription factors at specific endogenous gene loci in vivo are providing new views of transcription mechanisms and regulation. One, currently specific to Drosophila, makes use of state-of-the-art optics combined with the natural amplification of signals in tissue containing polytene chromosomes, allowing investigation of molecular interactions and dynamics at specific loci in real time in living nuclei3. The other, although having a history in Drosophila4,5,6, is species-general and uses crosslinking _in vivo_—that is, chromatin immunoprecipitation (ChIP) assays and variants thereof—to produce a ‘molecular image’ of specific protein interactions and chromatin modifications with particular DNA sequences in the genome7. Here I describe how these optical and molecular imaging approaches are providing new insights into transcription and the rate-limiting steps in its regulation in multicellular organisms. In particular, recent genome-wide ChIP assays in Drosophila8,9 and mammals10 support a paradigm shift in gene regulation by indicating that control of transcription elongation by RNA polymerase II (Pol II) in a promoter-proximal pausing model (Box 1), rather than control of the recruitment or initiation of Pol II, is the rate-limiting step for a large fraction of highly regulated genes.
Early insights from spread polytene nuclei
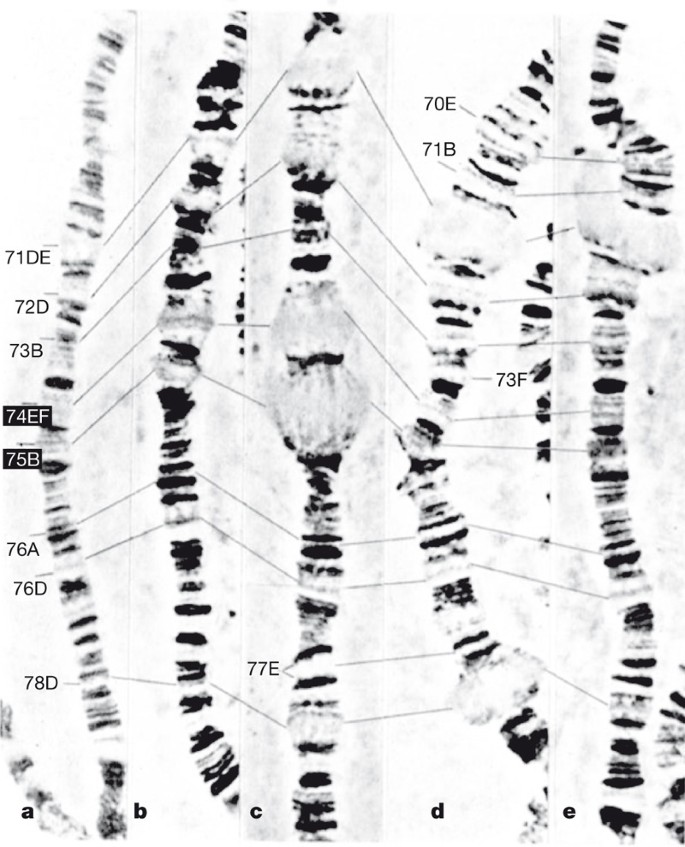
Over the past several decades, Drosophila has provided extraordinary views of chromosome structure and gene regulation. Simple phase microscopy of fixed and spread polytene nuclei provided early Drosophila geneticists with a high-resolution view of interphase chromosomes and a physical framework for their genetic maps11. This allowed genes to be positioned relative to the characteristic band and interband patterns seen along the lengths of each chromosome (Fig. 1). Particularly striking are the changes in polytene chromosome structure that occur when genes become transcriptionally activated12. High levels of transcription activation result in the decondensation of chromatin at specific genetic loci to form interbands and, in some cases, large distended chromosome puffs. These puffs provide cytological landmarks along each chromosome that come and go during the course of development, with each new set of puffs appearing, and old puffs disappearing, in response to waves of activation and repression of gene transcription (Fig. 1)12.
Figure 1: Puffing patterns on chromosome 3L during Drosophila larval development.
The chromosomes in a–e were isolated from progressively later developmental stages of third instar larvae. The light-staining distended puffs represent regions of high transcription activity. As developmental genes are turned on and off, the puffs appear and disappear. Reproduced with kind permission of Springer Science and Business Media from ref. 12.
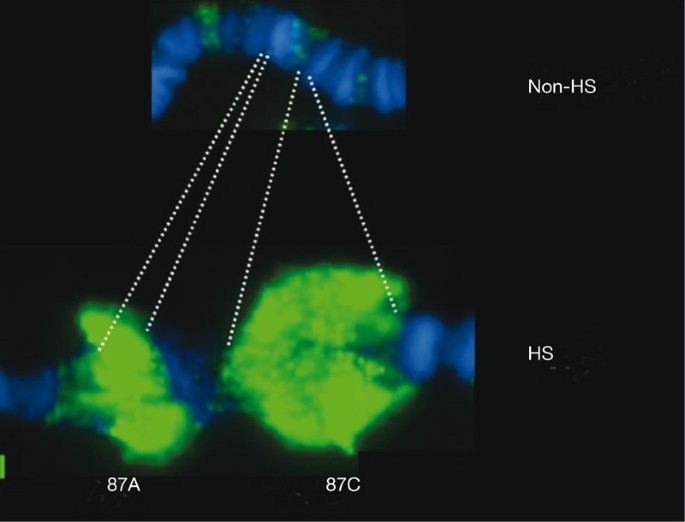
Environmental signals also activate the transcription of sets of genes, the activity of which can be observed as chromosomal puffs. The initial discovery in 1962 of the rapidly and highly activated heat shock response was not made by biochemists observing the induction of new messenger RNAs and proteins, but rather by a cytologist who was impressed by a striking new puffing pattern13. These observations of dramatic changes in cytology (Fig. 2) foreshadowed the changes in the molecular composition and dynamics of genes that would be uncovered by many laboratories in the decades that followed2.
Figure 2: Heat-shock-induced puffing at major heat shock loci 87A and C.
Displayed is a small segment of fixed chromosome 3 before (top) and after (bottom) heat shock. Chromosomes are stained for DNA (Hoeschst dye; blue) and for Pol II (green)29. HS, heat shock.
The genes that underlie these heat-shock-activated chromosomal puffs form a frequently used model for investigating mechanisms of gene activation and the accompanying changes in chromatin and chromosome structure. The staining of polytene chromosomes with antibodies to specific transcription factors14 allowed these factors to be visualized at these readily inducible loci and, more globally, over the entire genome. The molecular composition and chromatin architecture of these loci, and the changes triggered by the activation of these genes, have often proven to be general features of other developmentally activated loci15.
Live-cell imaging during gene activation
The ability to attach green fluorescent protein (GFP) and related fluorescent tags on chromosomal proteins has added a vital temporal dimension to the analysis of protein–DNA and protein–protein interactions in nuclei and on chromosomes. Expression of GFP-tagged proteins in mammalian cell lines allows the dynamics of transcription factors and chromatin components to be examined in nuclei in real time16,17. Fluorescence recovery after photobleaching (FRAP) experiments additionally reveal the dynamics of a nuclear protein within an arbitrarily chosen nuclear region18, or at a specific transgenic locus containing tandemly repeated genes16. Examination of specific, native gene loci requires more sensitivity and higher-resolution views of interphase nuclei. The giant interphase nuclei of Drosophila polytene tissue provide both the sensitivity and effective resolution to detect signals from specific chromosomal loci3.
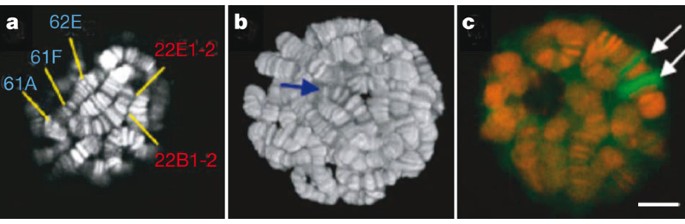
The advent of new optical-sectioning capabilities added further power to the imaging of intact nuclei. Computational deconvolution methods were used initially with wide-field microscopy to optically section Drosophila polytene nuclei19. Confocal, spinning disk and multi-photon microscopy further enhanced sectioning capabilities, with multi-photon microscopy being particularly effective in minimizing photodamage and providing high effective-resolution of thick biological specimens3. By applying these approaches to Drosophila polytene nuclei, it is possible to observe protein interactions with specific endogenous genetic loci in real time (Fig. 3). This was invaluable in directly tracking heat shock transcriptional activator (HSF) movement from nucleoplasm to specific heat shock gene loci (Hsp70), and for demonstrating that activated HSF remains bound in a non-exchanging state for many dozens of cycles of transcription3. These optical strategies should prove to be critical in exploring challenging mechanistic questions concerning transcription regulation, as discussed later.
Figure 3: Two-photon image of living salivary gland nuclei.
DAPI (4,6-diamidino-2-phenylindole)-stained DNA of a single nuclear section (a), a three-dimensional reconstructed nucleus (b), and GFP–Pol II at heat shock puffs (c; Pol II, green)3.
Molecular imaging of proteins on genes in vivo
As a complement to the microscopic views of nuclear protein distributions on polytene chromosomes, molecular approaches using protein–DNA crosslinking and immunoprecipitation of protein–DNA complexes (ultraviolet (UV)-ChIP and formaldehyde-ChIP) were developed in the mid-1980s to provide higher-resolution images of the molecular architecture of proteins and genes in vivo4,5,6. Applying these methods both before and during the time course of gene activation identified the timing and location of numerous protein interactions at high resolution (∼200 base pairs). The immediate, synchronous and robust heat shock response allows the heat shock genes to be examined before, and in the seconds that follow, stimulation, such that changes in chromatin and factor recruitment can be followed as a wave that passes along the length of these transcription units20. Additionally, the role of particular factors in these processes can be readily assessed using existing mutants or inhibitory drugs or by RNA interference knock-down strategies2.
An early and surprising finding, seen initially by UV-ChIP21, was that Drosophila heat shock genes had a polymerase associated with their promoters before activation, and that this associated Pol II was engaged in transcription and competent for elongation as determined by nuclear run-on assays (Box 1)22. These findings seemed to contradict the widely held view that the recruitment of Pol II or the initiation of transcription was the rate-limiting step in gene activation. Additional support for the existence and characterization of the nature of this Pol II complex came from the mapping of promoter melting with KMnO4 (ref. 23), which revealed melted DNA in the regions of heat shock genes residing 20–50 base pairs downstream of the transcription start sites. Also, the isolation and sizing of the chain-terminated run-on transcripts24 provided near-nucleotide-resolution mapping of pause sites to this same region, as two peaks separated by 10 base pairs. Interestingly, the RNAs are progressively more 5′-capped as they progress through the pause region, suggesting that capping occurs as soon as the RNA emerges from Pol II.
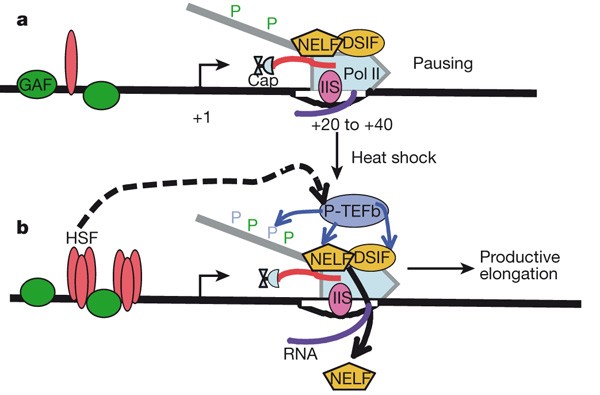
Two protein complexes—DRB sensitivity-inducing factor (DSIF; made up of spt4 and Spt5) and negative elongation factor (NELF; made up of five subunits)—were found to cooperate to repress transcription elongation in vitro25, and their negative effects could be overcome by P-TEFb (made up of Cdk9 and CycT) kinase26. Using ChIP assays, both DSIF and NELF were found to be located along with paused Pol II in the promoter-proximal regions of uninduced heat shock genes (Fig. 4)2,27. The P-TEFb kinase is recruited to active genes where it overcomes the negative effects through its kinase activity, which can phosphorylate DSIF, NELF and Ser 2 residues of the carboxy-terminal domain of the largest subunit of Pol II2,26. Interestingly, P-TEFb recruitment to heat shock genes depends on the heat shock activator HSF, but HSF does not seem to bind directly to P-TEFb15. Some, but not all, activators have been shown to interact with P-TEFb, so other mechanisms of P-TEFb recruitment need to be considered26. The transition into productive elongation seems to correlate with the loss of NELF from the promoter27. In contrast, DSIF remains associated with productively elongating Pol II and is thought to have a positive role after escape from the pause2,28. Paused polymerases are susceptible to backtracking, and the presence of elongation factor TFIIS at the pause region in vivo in Drosophila may stimulate the intrinsic RNA cleavage activity of Pol II to create a new RNA 3′ end in the active site, and thereby maintain a population of elongationally competent complexes29.
Figure 4: Minimal model of Pol II pausing at Hsp70 genes before and after heat shock.
For a more complete description of factors associated with heat shock genes see the more comprehensive review2. a, Prior to heat-shock, paused Pol II, which is partially phosphorylated at Ser 5 residues (green P) of the carboxy-terminal domain, is in a complex with DSIF and NELF complexes and occupies a region between 20–40 base pairs downstream of the start site of Hsp70. GAF is a sequence-specific binding factor that is present before activation. b, HSF is the key activator protein that trimerizes and binds with high affinity to its DNA elements in response to heat shock. Both DSIF and TFIIS (IIS) are part of both the paused and the fully competent Pol II elongation complexes. P-TEFb is the kinase that is critical for the maturation of paused Pol II into a productive elongation product, and it phosphorylates DSIF, NELF and the Ser 2 residues (blue P) of the carboxy-terminal domain of Pol II26.
Defining a Pol II as ‘promoter paused’
The presence of a peak of Pol II at the promoter is not sufficient to identify a Pol II as paused. Although such a distribution is indicative of Pol II promoter recruitment not being rate limiting in transcription, this Pol II could be in a pre-initiation complex or at some other post-recruitment step that precedes pausing. Additional criteria need to be applied30. Nuclear run-on assays can demonstrate that the detected Pol II is trancriptionally engaged, particularly if performed in the presence of Sarkosyl or high salt, which blocks new initiation and seems to remove barriers to elongation22. The isolation and sizing of intentionally terminated run-on RNAs show, at near-nucleotide-resolution, the location of the paused RNAs in vivo, and have the additional benefit of determining the capping signature, which in the cases examined is absent from the earliest pause sites, but is nearly complete by position +30 (ref. 24). Promoter-melting assays using KMnO4 can be performed on intact cells for short periods (30 s) to provide a snapshot of the reactivity of T residues, the hyper-reactivity of which is indicative of Pol II-melted DNA23. Whereas these melted residues tend to cluster in the region of 20–50 base pairs and highly correlate with paused Pol II, the pattern of reactivity can be influenced by other proteins either protecting or altering the reactivity of T residues. Additional signatures can be detected by ChIP assays and include the presence of NELF, and Ser 5- but not Ser 2-phosphorylated carboxy-terminal domain of the largest subunit (RpII215) of Pol II20,27.
How general is paused Pol II?
Pol II paused on promoters was regarded initially as a feature of the heat shock genes and a few other rapidly responsive genes. A small set of randomly chosen Drosophila genes also seemed to have paused Pol II, as assessed by UV-ChIP and nuclear run-on assays31, though not at the full occupancy (1 Pol II per promoter) seen for Hsp70 (ref. 30). Additionally, evidence supporting some form of elongational control in specific vertebrate genes has existed since the early 1980s, for example, greater nuclear run-on signals from 5′ portions than 3′ regions of chicken β-globin32. Similar data for mammalian c_-Myc_ and c_-Fos_ were augmented with in vitro studies that suggested initially a termination control further downstream, but more thorough characterizations in vivo showed these genes to have properties like those of Drosophila Hsp70 paused Pol II33,34.
More recently, genome-wide ChIP analyses of Pol II in human cells showed Pol II peaks in the promoter regions of a large fraction of genes35, and a study in human stem cells and differentiated cells showed that the majority of genes have peaks of Pol II that seem to have undergone transcription initiation, on the basis of their pattern of histone H3K4 trimethylation (a modification linked to active promoters) and the production of short 5′ transcripts10. Interestingly, many of these genes are not producing mature transcripts1,10. Very recent genome-wide studies in Drosophila have revealed that many genes have peaks of paused Pol II, including important regulatory genes of early Drosophila development8,9. Numbers of promoters with paused Pol II in Drosophila, as defined by exhibiting additional signatures of paused Pol II such as NELF occupancy and promoter melting8,9, are estimated conservatively to be about 20% of all genes that have any associated Pol II.
Is this mechanism of elongation control used only for the estimated 20% of genes in Drosophila7,8, and >50% (estimated by different criteria) in human stem cells, or is pausing a still more universal step on the pathway of transcription of most genes? The inhibition of P-TEFb kinase, (a kinase which is critical for Pol II escape from pausing into productive elongation), can lead to an 80% reduction of Pol II transcription36, indicating that these steps may be pervasive, though not necessarily rate limiting, under the particular cellular conditions examined.
Questions and future prospects
What are the proteins and RNAs that set up and maintain paused Pol II, and what is the mechanism used to achieve this? In Drosophila, GAGA factor (GAF; also known as Trl) is bound before heat shock induction and seems to be critical for setting up the pause on Hsp70 (refs 37 and 38). However, many, but not all, genes showing paused Pol II have associated GAF, and GAF homologues are not obvious in mammals. Is there a class of transcription factors with GAF-like function, and how do they function mechanistically? Although NELF and DSIF are major players, their depletion leads to neither a complete loss of paused Pol II nor high constitutive expression. Do other factors, and perhaps inherent features of Pol II and the underlying DNA or RNA sequence, also participate in this process? Can we devise other genome-wide approaches that allow characterization of paused Pol II RNAs at nucleotide-resolution and thereby improve the search for global sequence elements?
How dynamic is the promoter-proximal paused Pol II? The paused Pol II has been described by some as performing abortive transcription, meaning that Pol II undergoes termination before completing a full-length transcript. An extreme version of this view posits that the paused Pol II is terminated and replaced with productive Pol II following gene activation. However, there is no evidence in vivo that abortive transcription is a necessary property of these paused Pol II molecules. Clearly, there are cases in which Pol II does abort transcription early in the process of transcription in eukaryotes, for example, HIV transcription. However, these short HIV transcripts are terminated downstream of the HIV pause region and these termination events are dictated by mechanisms distinct from pausing39. In the case of Drosophila Hsp70, short transcripts do not accumulate, although some fraction of transcripts fail to efficiently elongate in run-on assays and are likely to be back-tracked and arrested. However, TFIIS is also present at the promoter and may allow these arrested Pol II complexes to be in dynamic equilibrium with elongationally competent paused complexes2,29. For some genes, Pol II shows only fractional promoter occupancy—less than one Pol II per promoter. Does that reflect only a fraction of the cells having a poised promoter and thus an active or activatable gene? Or does this variation in occupancy reflect differences in the relative rates with which Pol II enters and escapes from the pause region? Both of these steps may be influenced by the spectrum of regulatory proteins bound to enhancer and promoter regions. These difficult questions concerning molecular dynamics at native gene loci should be addressable with the improved optical approaches discussed above, and with carefully developed in vitro systems that recapitulate all of the in vivo landmarks of these Pol II complexes.
How do activator proteins interface with paused Pol II to influence its escape to productive elongation? Clearly the P-TEFb kinase is a critical executer of the activation signal, and its presence and ability to phosphorylate components of the paused Pol II complex are critical for stimulating productive elongation26. In some cases, activators are known to interact with P-TEFb, at least in simple pull-down or co-immunoprecipitation assays26; however, in other cases, activators show no detectable affinity for P-TEFb15, and other existing mechanisms26, or novel mechanisms yet to be discovered, allow activators to communicate with P-TEFb.
In summary, the pervasiveness of paused Pol II is causing the re-evaluation of the long-held textbook view of transcription and its regulation. For decades, the major mechanism of gene regulation in higher eukaryotes was thought to reside at the level of either recruitment of Pol II to promoters or transcription initiation. In light of genome-wide data7,8,9,10, a post-recruitment and early elongation mechanism needs to be considered as a major mode of regulation in higher eukaryotes. Drosophila has been a useful model system for past characterization of this regulation and its unique and shared features hold great promise for its role in addressing these outstanding questions.
References
- ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816 (2007)
- Saunders, A., Core, L. J. & Lis, J. T. Breaking barriers to transcription elongation. Nature Rev. Mol. Cell Biol. 7, 557–567 (2006)
Article CAS Google Scholar - Yao, J., Munson, K. M., Webb, W. W. & Lis, J. T. Dynamics of heat shock factor association with native gene loci in living cells. Nature 442, 1050–1053 (2006)
Article CAS ADS Google Scholar - Gilmour, D. S. & Lis, J. T. In vivo interactions of RNA polymerase II with genes of Drosophila melanogaster . Mol. Cell. Biol. 5, 2009–2018 (1985)
Article CAS Google Scholar - Solomon, M. J., Larsen, P. L. & Varshavsky, A. Mapping protein–DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell 53, 937–947 (1988)
Article CAS Google Scholar - Orlando, V. & Paro, R. Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell 75, 1187–1198 (1993)
Article CAS Google Scholar - Ren, B. et al. Genome-wide location and function of DNA binding proteins. Science 290, 2306–2309 (2000)
Article CAS ADS Google Scholar - Zeitlinger, J. et al. A polymerase stalling at developmental control genes in the Drosophila embryo. Nature Genet. (in the press)
- Muse et al. RNA polymerase is poised for activation across the genome. Nature Genet. (in the press)
- Guenther, M. G., Levine, S. S., Boyer, L. A., Jaenisch, R. & Young, R. A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130, 77–88 (2007)
Article CAS Google Scholar - Bridges, C. B. Salivary chromosome maps with a key to the banding of the chromosome of Drosophila melanogaster . J. Hered. 26, 60–64 (1935)
Article Google Scholar - Ashburner, M. in Developmental studies on giant chromosomes, (ed. W. Beermann) 101–151 (Springer, New York, 1972)
Book Google Scholar - Ritossa, F. A new puffing pattern induced by temperature shock and DNP in Drosophila . Experientia 18, 571–573 (1962)
Article CAS Google Scholar - Silver, L. M. & Elgin, S. C. Production and characterization of antisera against three individual NHC proteins; a case of a generally distributed NHC protein. Chromosoma 68, 101–114 (1978)
Article CAS Google Scholar - Lis, J. T., Mason, P., Peng, J., Price, D. H. & Werner, J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 14, 792–803 (2000)
CAS PubMed PubMed Central Google Scholar - Hager, G. L., Elbi, C. & Becker, M. Protein dynamics in the nuclear compartment. Curr. Opin. Genet. Dev. 12, 137–141 (2002)
Article CAS Google Scholar - Janicki, S. M. et al. From silencing to gene expression: real-time analysis in single cells. Cell 116, 683–698 (2004)
Article CAS Google Scholar - Kimura, H., Sugaya, K. & Cook, P. R. The transcription cycle of RNA polymerase II in living cells. J. Cell Biol. 159, 777–782 (2002)
Article CAS Google Scholar - Mathog, D., Hochstrasser, M., Gruenbaum, Y., Saumweber, H. & Sedat, J. Characteristic folding pattern of polytene chromosomes in Drosophila salivary gland nuclei. Nature 308, 414–421 (1984)
Article CAS ADS Google Scholar - Boehm, A. K., Saunders, A., Werner, J. & Lis, J. T. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol. Cell. Biol. 23, 7628–7637 (2003)
Article CAS Google Scholar - Gilmour, D. S. & Lis, J. T. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol. Cell. Biol. 6, 3984–3989 (1986)
Article CAS Google Scholar - Rougvie, A. E. & Lis, J. T. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54, 795–804 (1988)
Article CAS Google Scholar - Giardina, C., Perez-Riba, M. & Lis, J. T. Promoter melting and TFIID complexes on Drosophila genes in vivo. . Genes Dev. 6, 2190–2200 (1992)
Article CAS Google Scholar - Rasmussen, E. B. & Lis, J. T. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc. Natl Acad. Sci. USA 90, 7923–7927 (1993)
Article CAS ADS Google Scholar - Yamaguchi, Y. et al. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97, 41–51 (1999)
Article CAS Google Scholar - Peterlin, B. M. & Price, D. H. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23, 297–305 (2006)
Article CAS Google Scholar - Wu, C. H. et al. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila . Genes Dev. 17, 1402–1414 (2003)
Article CAS Google Scholar - Zhu, W. et al. DSIF contributes to transcriptional activation by DNA-binding activators by preventing pausing during transcription elongation. Nucleic Acids Res. 35, 4064–4075 (2007)
Article CAS Google Scholar - Adelman, K. et al. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol. Cell 17, 103–112 (2005)
Article CAS Google Scholar - Lis, J. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb. Symp. Quant. Biol. 63, 347–356 (1998)
Article CAS Google Scholar - Rougvie, A. E. & Lis, J. T. Postinitiation transcriptional control in Drosophila melanogaster . Mol. Cell. Biol. 10, 6041–6045 (1990)
Article CAS Google Scholar - Gariglio, P., Bellard, M. & Chambon, P. Clustering of RNA polymerase B molecules in the 5′ moiety of the adult _β_-globin gene of hen erythrocytes. Nucleic Acids Res. 9, 2589–2598 (1981)
Article CAS Google Scholar - Krumm, A., Meulia, T., Brunvand, M. & Groudine, M. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 6, 2201–2213 (1992)
Article CAS Google Scholar - Albert, T. et al. Nucleosomal structures of c-myc promoters with transcriptionally engaged RNA polymerase II. Mol. Cell. Biol. 17, 4363–4371 (1997)
Article CAS Google Scholar - Kim, T. H. et al. A high-resolution map of active promoters in the human genome. Nature 436, 876–880 (2005)
Article CAS ADS Google Scholar - Chao, S. H. & Price, D. H. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo . J. Biol. Chem. 276, 31793–31799 (2001)
Article CAS Google Scholar - Lee, H., Kraus, K. W., Wolfner, M. F. & Lis, J. T. DNA sequence requirements for generating paused polymerase at the start of hsp70 . Genes Dev. 6, 284–295 (1992)
Article CAS Google Scholar - Wang, Y. V., Tang, H. & Gilmour, D. S. Identification in vivo of different rate-limiting steps associated with transcriptional activators in the presence and absence of a GAGA element. Mol. Cell. Biol. 25, 3543–3552 (2005)
Article CAS Google Scholar - Henderson, A. J. & Gilmour, D. S. Transcription termination factor Pcf11 limits the processivity of Pol II on an HIV provirus to repress gene expression. Genes Dev. 21, 1609–1614 (2007)
Article Google Scholar - Ring, B. Z., Yarnell, W. S. & Roberts, J. W. Function of E. coli RNA polymerase sigma factor sigma 70 in promoter-proximal pausing. Cell 86, 485–493 (1996)
Article CAS Google Scholar
Acknowledgements
I thank J. Werner for generating the immunofluorescence images used in Fig. 2, and M. Hamblin, D. Gilmour, K. Adelman and my laboratory members for helpful comments on the manuscript. This work was supported by an NIH grant.
Author information
Author notes
- Correspondence and requests for materials should be addressed to D.L. (jtl10@cornell.edu).
Authors and Affiliations
- Molecular Biology and Genetics, 416 Biotechnology Building, Cornell University, Ithaca, New York 14853, USA ,
John T. Lis
Authors
- John T. Lis
You can also search for this author inPubMed Google Scholar
Ethics declarations
Competing interests
The author declares no competing financial interests.
Rights and permissions
About this article
Cite this article
Lis, J. Imaging Drosophila gene activation and polymerase pausing in vivo.Nature 450, 198–202 (2007). https://doi.org/10.1038/nature06324
- Received: 06 September 2007
- Accepted: 28 September 2007
- Issue Date: 08 November 2007
- DOI: https://doi.org/10.1038/nature06324