Proportionally more deleterious genetic variation in European than in African populations (original) (raw)
Main
Current estimates of the number of deleterious mutations per diploid human genome vary by several orders of magnitude. Using a correlation in inbreeding rates within consanguineous marriages and mortality, Morton et al.5 estimated that each of us carries three to five lethal equivalents (that is, an allele or combination of alleles that if made homozygous would be lethal), whereas Kondrashov6 has predicted that the number may be as high as 100 lethal equivalents. Comparative genomic methods indicate that about 38% of amino-acid-changing polymorphisms are deleterious, with 1.6 new deleterious mutations arising per individual per generation7, whereas studies based on segregating polymorphisms estimate that each person carries between 500 and 1,200 deleterious mutations3,8. It is difficult to reconcile these estimates because each study used different methods and data. Furthermore, studies that used DNA sequences included data from only several hundred genes. Hence there is a crucial need for an unbiased genome-wide estimate of the number of damaging mutations carried by individuals in different populations.
We quantify the number of damaging mutations per diploid human genome by combining the Applera genome-wide survey of SNPs found by the resequencing of 20 EAs and 15 AAs9 with comparative genomic data including the PanTro2 build of the chimpanzee genome and predictions from protein structures. After applying strict quality control criteria, the data set that we analysed contains 39,440 autosomal SNPs free of ascertainment bias, comprising 10,150 unique transcripts in the human genome (see Methods). Of these SNPs, 20,893 were synonymous (nucleotide changes that do not change the amino acid) and 18,547 were non-synonymous (nucleotide changes that change the amino acid).
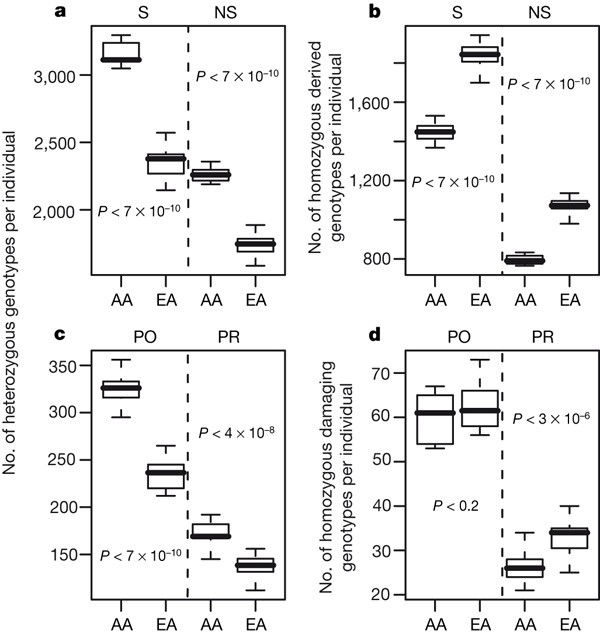
At each SNP, an individual can be homozygous for the ancestral allele (carrying zero copies of the mutant allele), heterozygous (carrying one copy of the mutant allele) or homozygous for the derived allele (carrying two copies of the mutant allele). We find that an individual is heterozygous, on average, for 1,962.4 ± 275.1 (mean ± s.d.) non-synonymous SNPs (Fig. 1a and Supplementary Table 1). These numbers are an underestimate because only SNPs with high-quality sequence and a matching chimpanzee base are considered. Perhaps for these reasons, our estimate is slightly smaller than that by Cargill et al.10, even after adjusting their estimate to account for the current estimated number of genes in the genome. For both synonymous and non-synonymous SNPs, AA individuals are heterozygous at a greater number of SNPs than EA individuals are (Fig. 1a; P < 6.2 × 10-10, Mann–Whitney _U_-test for synonymous SNPs; P < 6.2 × 10-10, Mann–Whitney _U_-test for non-synonymous SNPs), which is consistent with previous studies finding higher levels of genetic variability in Africa4. For both types of SNP we find that EA individuals are homozygous for the derived allele at a greater number of SNPs than AA individuals are (Fig. 1b; P < 6.2 × 10-10, Mann–Whitney _U_-test). These patterns are due largely to a greater number of SNPs fixed for the derived allele in the EA sample while segregating for two alleles in the AA sample, because when we count the number of homozygous derived genotypes per individual in the EA individuals considering only those SNPs that are polymorphic in EAs (that is, excluding SNPs that are fixed in EAs but polymorphic in AAs) and the number of homozygous derived genotypes per individual in the AA individuals considering only those SNPs polymorphic in AAs (for example excluding SNPs that are fixed in AAs and polymorphic in EAs), the EA individuals no longer have significantly more homozygous derived genotypes for both categories of SNP.
Figure 1: Distribution of the number of heterozygous and homozygous genotypes per individual.
a, Number of heterozygous genotypes per individual at synonymous (S) or non-synonymous (NS) SNPs. b, Number of genotypes homozygous for the derived allele per individual at synonymous or non-synonymous SNPs. c, Number of heterozygous genotypes per individual at possibly damaging (PO) or probably damaging (PR) SNPs. d, Number of genotypes homozygous for the damaging allele at possibly damaging or probably damaging SNPs. Dark horizontal lines within boxes indicate medians, and the whiskers indicate the ranges of the distributions.
To estimate the number of damaging alleles carried by each individual in our sample, we used the PolyPhen algorithm8,11 to predict which non-synonymous SNPs might disrupt protein function. PolyPhen predicts whether a SNP is ‘benign’, ‘possibly damaging’ or ‘probably damaging’ on the basis of evolutionary conservation and structural data. To assess whether ‘damaging’ SNPs were more likely to be deleterious, we compared the allele frequency distribution of SNPs predicted to be ‘benign’, ‘possibly damaging’ and ‘probably damaging’ for each population. We find that the three distributions are significantly different from each other, with more low-frequency SNPs in the ‘probably damaging’ category (Table 1; P < 5.9 × 10-81 for AA, P < 2.3 × 10-101 for EA; Kruskal–Wallis test), suggesting that most SNPs classified as ‘damaging’ are also evolutionarily deleterious.
Table 1 Distribution of Applera SNPs by population and functional class
Figure 1c, d shows the distribution of the number of SNPs per individual where individuals were heterozygous (Fig. 1c) and homozygous for the damaging allele (Fig. 1d) for SNPs predicted to be ‘possibly damaging’ and ‘probably damaging’. We find that an individual typically carries 426.1 damaging (here defined as ‘possibly damaging’ or ‘probably damaging’) SNPs in the heterozygous state (s.d. 65.4, range 340–534) and 91.7 in the homozygous state (s.d. 8.6, range 77–113). Because we surveyed just over 10,000 genes, the actual number of damaging mutations in a person’s genome may be as much as double that given here. Every individual in our sample is heterozygous at fewer ‘probably damaging’ SNPs than synonymous SNPs, which is consistent with the elimination of damaging SNPs from the population by purifying selection. AAs have significantly more heterozygous genotypes than EAs for all three PolyPhen categories (Fig. 1c; P < 6.2 × 10-10 for ‘possibly damaging’ SNPs; P < 3.7 × 10-8 for ‘probably damaging’ SNPs). The two populations differ significantly in the distribution of homozygous genotypes for the damaging allele at ‘probably damaging’ SNPs (Fig. 1d; P < 2.7 × 10-6), with EAs having about 26% more homozygous damaging genotypes than AAs. The lack of a statistical difference at ‘possibly damaging’ SNPs (P = 0.17) is probably due to a lack of power because, overall, all other categories of SNPs (synonymous, non-synonymous, ‘benign’ and ‘probably damaging’) follow the same pattern of excess homozygosity for the derived/damaging allele in EAs relative to AAs.
Classical analyses of human inbreeding indicate that each individual carries 1.44–5 lethal equivalents5,12. However, inbreeding studies cannot determine whether a single lethal equivalent is due to one lethal allele, two alleles each with a 50% chance of lethality, ten alleles each with a 10% chance of lethality, or other combinations. Because we find that individuals carry hundreds of damaging alleles, it is likely that each lethal equivalent consists of many weakly deleterious alleles. Our finding that each person carries several hundred potentially damaging SNPs indicates that large-scale medical resequencing will be useful to find common and rare SNPs of medical consequence2.
We next examined the distribution of synonymous and non-synonymous SNPs between AA and EA population samples (Table 1). As expected4, there are more of both types of SNP in the AA sample than in the EA sample. However, when classifying synonymous and non-synonymous SNPs as being shared, private to AAs or private to EAs, we strongly reject homogeneity (Table 2, P < 3.0 × 10-88). We find the proportion of private SNPs that are non-synonymous (49.9%) to be higher than the proportion of shared SNPs that are non-synonymous (41.7%; P < 4.3 × 10-54), which is not surprising because non-synonymous SNPs are more likely to be at a lower frequency and thus be population specific. However, considering only the private SNPs, we find that the EA sample has a higher proportion of non-synonymous SNPs (55.4%) than the AA sample (47.0%; P < 2.3 × 10-37). We observed a similar significant proportional excess of private non-synonymous SNPs in an independent data set collected by the SeattleSNPs project (Supplementary Table 3 and Supplementary Notes). The SeattleSNPs data, additional quality control analyses (Supplementary Table 4 and Supplementary Notes), and a similar finding reported for the ANGPTL4 locus13 indicate that this pattern is not an artefact of the Applera data. Our further analyses with Yoruban samples from Nigeria collected by the International HapMap Consortium14 support this result, indicating that it is robust to admixture (Supplementary Notes).
Table 2 Results of _G_-tests of homogeneity for Table 1
We propose that the proportional excess of non-synonymous polymorphism in the EA sample could be due to the varying efficacy of purifying selection resulting from differences in demographic histories between the two populations. Our hypothesis has two testable predictions: first, if this proportional excess of non-synonymous polymorphisms in EAs is due to an excess of damaging alleles, we would also expect to find a proportional increase of ‘probably damaging’ SNPs as predicted by PolyPhen in the EA sample; and second, we should be able to recapitulate this pattern by using simulations with reasonable demographic parameters. When dividing non-synonymous SNPs into the three PolyPhen categories, we find a significant excess of ‘probably damaging’ SNPs in private SNPs compared with shared SNPs (Tables 1 and 2). When considering only the private SNPs, we find a significantly higher proportion of ‘probably damaging’ SNPs in the EA sample relative to the AA sample (Tables 1 and 2; P < 3.3 × 10-11), supporting our hypothesis that the excess proportion of non-synonymous SNPs in the EA sample is due to a higher proportion of damaging SNPs.
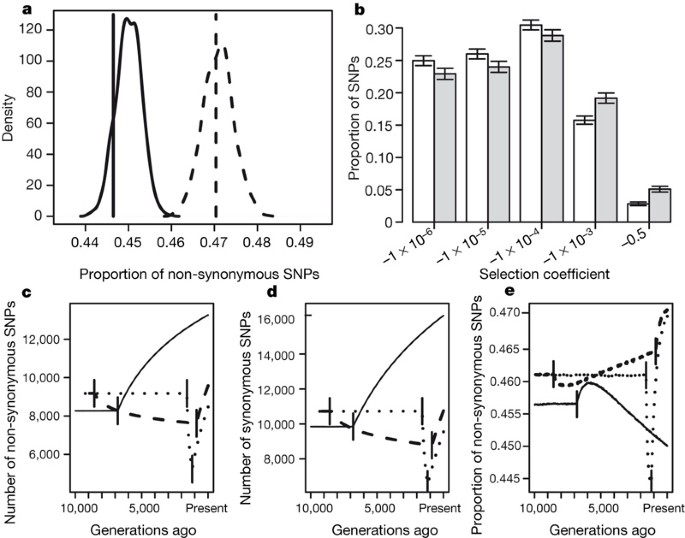
To assess whether these observations are consistent with plausible demographic histories of the two populations, we developed a large-scale forward simulation program that included non-stationary demography and a negative log-normal distribution of selective effects for deleterious mutations. Our program used demographic parameters estimated from the data and the literature15 for each population (Supplementary Table 2). For example, for the simulations in Fig. 2a, b we used a population expansion model for the AAs and a bottleneck model for the EAs (Supplementary Fig. 1). We sampled from these simulated populations and found that the proportion of non-synonymous SNPs is greater in the bottlenecked population than in a population that has expanded (Fig. 2a, Supplementary Table 2 and Supplementary Fig. 2a). Furthermore, as shown in Fig. 2a, the simulated proportions agree with the observed proportions for the Applera data set (here the proportion includes all SNPs, not just private ones). For all demographic models considered, we observed a higher proportion of non-synonymous SNPs in the population that underwent a bottleneck than in a population of constant size or in one that has expanded; however, the degree to which these other models fit the observed data is variable (Supplementary Table 2 and Supplementary Fig. 2a). For all models tested, we find that a higher proportion of SNPs in the simulated EA sample are weakly or strongly deleterious (-0.001 < s < -0.5) than in the simulated AA sample (Fig. 2b, Supplementary Table 2 and Supplementary Fig. 2b), which supports our hypothesis that a higher proportion of deleterious alleles have accumulated in the bottlenecked population. Our analysis illustrates that plausible models of human demography and purifying selection are sufficient to account for the observed increase in the proportion of non-synonymous SNPs in the EA sample relative to the AA sample.
Figure 2: Demography and selection can cause a proportional excess of non-synonymous SNPs in Europeans.
a, b, Results of forward simulations of a population that expanded (AA 2 in Supplementary Table 2), to represent the AA population and a population that experienced a bottleneck to represent the EA population (EA 1 in Supplementary Table 2). a, Distribution of the proportion of non-synonymous SNPs segregating in samples simulated under European (dashed curve) and African (solid curve) demographic models. Vertical lines show the observed proportions in the Applera data set. b, Distribution of selection coefficients for simulated SNPs in the AA (white bars) and the EA (grey bars) samples. The labels on the x axis are the more negative limits of the bins. Error bars denote 95% intervals on the proportion of SNPs in each group. c–e, Expected distribution of SNPs over time during a population expansion (AA 2, solid lines), a long, mild bottleneck (EA 1, dashed lines) and a short, severe bottleneck (EA 6, dotted lines). Time moves forward from left to right. Solid vertical lines indicate when the populations changed size. Further details are given in Supplementary Table 2. c, The number of non-synonymous SNPs. d, The number of synonymous SNPs. e, The proportion of non-synonymous SNPs.
To determine how the bottleneck contributed to the increased proportion of non-synonymous SNPs in the EA sample, we recorded the number of SNPs at different time points throughout our forward simulations (see Methods). Figure 2c–e shows how the number of synonymous SNPs and non-synonymous SNPs and the proportion of non-synonymous SNPs change over time for the EA and AA models described above as well as for a second bottleneck model having a shorter, but more severe, reduction in population size. At the start of the bottleneck, the proportion of non-synonymous SNPs drops below the pre-bottleneck value (because of the preferential loss of low-frequency non-synonymous SNPs). Then the proportion increases during the bottleneck as a result of the accumulation of slightly deleterious SNPs that behave almost neutrally in the small population but are eliminated efficiently from larger populations16. Once the population expands, the proportion of non-synonymous SNPs increases markedly because the increase in population size results in many more mutations (most of which are non-synonymous, because of the genetic code) entering the population (Fig. 2c, d). Because growth was recent, purifying selection has not had sufficient time to decrease the proportion of non-synonymous SNPs to the equilibrium value for the larger population. A related effect has been noted in spatial expansion models, in which deleterious mutations can ‘surf’ to high frequency on the edge of the expansion17. Our simulations for African demography suggest that once the African population expanded, the proportion of non-synonymous SNPs also increased initially. However, because the African expansion occurred farther back in time than the most recent European expansion, the proportion of non-synonymous SNPs has had more time to decrease closer to the equilibrium value in the AA sample. At the present time, the absolute numbers of SNPs are higher in the non-bottleneck model (AA 2) than in the bottleneck models (EA 1 and EA 6). The bottleneck dynamics were robust to the distribution of selective effects used in our simulations (Supplementary Fig. 3).
Thus, both the PolyPhen analysis and the forward simulations suggest that, given the lower levels of genetic diversity found in Europeans than in Africans, the former have a higher proportion of deleterious alleles, which can be explained by the ‘out of Africa’ bottleneck and subsequent expansion that outbred European populations endured. This result is important for two reasons. First, whereas previous work has highlighted examples of European-specific positive selection14,18,19,20,21, the importance of adaptations for the evolution of European populations needs to be tempered by our finding that negative selection is less effective at removing slightly deleterious alleles from European populations. Second, the idea that bottlenecks and founder effects could lead to an increase in damaging alleles in human populations was historically reserved for isolated populations that experienced severe founder effects (for example Ashkenazi Jews22 and Finns23). Our work suggests that the interaction of demographic processes and purifying selection can have an important impact on the distribution of deleterious variation, even in populations that did not undergo a severe founder effect.
Methods Summary
We used an improved bioinformatics pipeline to analyse SNPs, described in ref. 9. We mapped the SNPs to the RefSeq v18 gene model to determine whether they were synonymous or non-synonymous. Ancestral and derived states for each SNP were determined with the syntenic net alignments between hg18 and panTro2 (refs 24, 25). When counting the number of genotypes per individual, we added a correction for misidentification of the ancestral allele26. SNPs were dropped from the analysis if they failed to meet our bioinformatics quality controls, but we did not filter SNPs on the basis of frequency.
To predict whether a non-synonymous SNP will damage protein function, we used an updated version of PolyPhen that has false-positive and false-negative rates below about 15% (Supplementary Methods). When counting the number of damaging genotypes per individual, we used the subset of SNPs in which the predicted damaging allele was the derived allele.
An additional four AA individuals were sequenced, but we did not include them (or SNPs private to them) in further analyses because we determined that they had substantially more European admixture than the other AAs (Supplementary Methods, Supplementary Table 5 and Supplementary Fig. 4). If our estimates of admixture are not perfect, this should not drastically affect the comparisons of different classes of SNPs, making our analysis robust to this problem (Supplementary Notes). The Coriell sample numbers for the individuals used in our study are given in Supplementary Table 1.
To test whether the higher proportion of non-synonymous SNPs in EAs than in AAs could be due to the different demographic histories of the two populations, we used forward simulations that allowed us to model demography and purifying selection. We considered a range of demographic models for both populations (Supplementary Table 2) and a distribution of selective effects for non-synonymous SNPs.
Online Methods
Bioinformatic pipeline
SNPs were mapped onto RefSeq v18 gene model in a two step process. First we aligned the Celera gene models to hg18 by using Blat v33.2 (ref. 27), filtering out any hits that had less than 98.5% sequence identity or less than 90% coverage. We then aligned RefSeq v18 CDS sequences28 to hg18 by using the same filtering conditions. Having coordinates of both our SNPs and RefSeq gene models relative to the assembly, we converted our SNP positions onto the RefSeq CDS position to determine the reading frame. If a SNP mapped to multiple RefSeqs, we chose the longest transcript for analysis. Any sequences in RefSeq that were not covered by PCR amplicons were excluded from analysis. SNPs that mapped to multiple RefSeqs that were out-of-frame were discarded. SNPs were polarized by the chimpanzee genome by using the syntenic net alignments between hg18 and panTro2 (refs 24, 25). SNPs were dropped from the analysis if they aligned to a non-syntenic region in panTro2, neither human allele matched the panTro2 allele, fewer than nine individuals in either population had successfully called genotypes, or if we detected a departure from Hardy–Weinberg equilibrium (defined as P < 0.01) with the exact test of Wigginton et al.29. SNPs mapping to multiple transcripts were counted only once. We used all SNPs passing bioinformatics quality controls, without filtering for frequency. Certain analyses were also performed excluding singletons and are described in Supplementary Notes.
Correction for ancestral misidentification
Misidentifying the ancestral state of a SNP can lead to miscalculating the proportion of homozygous derived SNPs carried by each individual. We accounted for the probability of ancestral misidentification by adapting the method of ref. 26 to model the number of homozygous SNPs carried by each individual. In this model, the number of homozygous SNPs carried by each individual is considered to be a mixture of sites whose ancestral states were correctly identified by using the chimpanzee outgroup and those that were not (two unknown quantities). The corrected number of homozygous derived mutations carried by each individual can then be reconstituted by solving for this unknown quantity as a function of the mixture proportions and observed data. Here, the mixture proportions account for the divergence time between human and chimpanzee by using a context-dependent mutation model inferred along the human lineage30.
PolyPhen analysis
We predicted the functional consequences of SNPs using a newer version of PolyPhen that differs slightly from that described in refs 8, 11. For SNPs mapping to multiple transcripts, we ran PolyPhen on the SNP in each transcript. If a SNP had different PolyPhen predictions in different transcripts, it was excluded from any further PolyPhen analyses; 340 SNPs had multiple PolyPhen predictions and 56 did not have a prediction. For our data, PolyPhen used 18.2 ± 28.0 (mean ± s.d.) sequences across covered SNPs. SNPs used for analyses, along with their frequencies and PolyPhen predictions, are available in Supplementary Information. For about 77.5% of the ‘benign’ SNPs, 98.2% of the ‘possibly damaging’ SNPs and 98.8% of the ‘probably damaging’ SNPs, the damaging allele (the allele with the lower PSIC (position-specific independent count) score) is the derived allele, indicating that PolyPhen has a greater ability to distinguish which allele is damaging for ‘probably damaging’ SNPs than for ‘benign’ or ‘possibly damaging’ SNPs. As explained in Supplementary Methods, PolyPhen classified 85.5% of 3,604 disease mutations annotated in the UniProt database as either ‘probably damaging’ or ‘possibly damaging’, while predicting 86.1% of 12,237 amino acid differences between humans and another mammalian orthologue as ‘benign’. These results suggest that the false positive and false negative rates of the algorithm are each below about 15%.
Counting the number of genotypes per individual
To determine whether AA individuals were heterozygous at more SNPs than EA individuals were, we used a two-sided Mann–Whitney _U_-test to compare the distribution of the number of heterozygous genotypes per individual in AA individuals with the distribution of the number of heterozygous genotypes per individual in the EA individuals. This comparison was done separately for synonymous, non-synonymous, ‘benign’, ‘possibly damaging’ and ‘probably damaging’ SNPs. A similar test was used to test whether EA individuals were homozygous for the derived allele at a greater number of SNPs than AA individuals. When counting the number of SNPs per individual, we wanted to ensure that our counts were not biased because some samples had more complete sequencing than others. We divided the number of genotypes in an individual of each particular category (for example the number of heterozygous genotypes for synonymous sites in a particular individual) by the total number of genotypes in that category (for example the total number of genotypes at synonymous sites) in the individual. We then tested whether the distribution of these proportions was different between the AA and EA samples. In all cases we observed the same pattern as that shown in Fig. 1 (data not shown), indicating that this result was not due to inconsistent sequencing of different individuals.
Forward simulations
A detailed description of the methods used for forward simulations is given in Supplementary Methods. In brief, we wanted to test whether the observation of a higher proportion of non-synonymous SNPs in EAs than in AAs could have been due to the different demographic histories of the two populations. We simulated one population forwards in time with a demographic history consistent with that of Africans and another population forwards in time with demographic history consistent with that of Western Europe. We considered a variety of plausible demographic models for each population15 and simulated the African and European populations independently of each other. In addition to simulating populations in which all SNPs were neutral, we also independently simulated a second set of populations for each set of demographic parameters in which the selection coefficients were from a distribution of selective effects (Supplementary Methods) to mimic non-synonymous sites. At the end of the simulation, we sampled 15 individuals from the population that expanded and 20 individuals from the population that underwent a bottleneck. We examined whether one population had a higher proportion of damaging (non-synonymous) SNPs and whether segregating SNPs in one population had a different distribution of selection coefficients than SNPs segregating in the other population.
Accession codes
Data deposits
Accession numbers for the SNP markers analysed in this study are dbSNP numbers ss48401226–ss48429818 and ss48429821–ss48431291, submitted under the handle APPLERA_GI, and ss86236910–ss86273113, submitted under the handle CORNELL.
References
- Muller, H. J. Our load of mutations. Am. J. Hum. Genet. 2, 111–176 (1950)
CAS PubMed PubMed Central Google Scholar - Cohen, J. C. et al. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science 305, 869–872 (2004)
Article CAS ADS PubMed Google Scholar - Fay, J. C., Wyckoff, G. J. & Wu, C. I. Positive and negative selection on the human genome. Genetics 158, 1227–1234 (2001)
CAS PubMed PubMed Central Google Scholar - Tishkoff, S. A. & Williams, S. M. Genetic analysis of African populations: human evolution and complex disease. Nature Rev. Genet. 3, 611–621 (2002)
Article CAS PubMed Google Scholar - Morton, N. E., Crow, J. F. & Muller, H. J. An estimate of the mutations damage in man from data on consanguineous marriages. Proc. Natl Acad. Sci. USA 42, 855–863 (1956)
Article CAS ADS PubMed PubMed Central Google Scholar - Kondrashov, A. S. Contamination of the genome by very slightly deleterious mutations: why have we not died 100 times over? J. Theor. Biol. 175, 583–594 (1995)
Article CAS PubMed Google Scholar - Eyre-Walker, A. & Keightley, P. D. High genomic deleterious mutation rates in hominids. Nature 397, 344–347 (1999)
Article CAS ADS PubMed Google Scholar - Sunyaev, S. et al. Prediction of deleterious human alleles. Hum. Mol. Genet. 10, 591–597 (2001)
Article CAS PubMed Google Scholar - Bustamante, C. D. et al. Natural selection on protein-coding genes in the human genome. Nature 437, 1153–1157 (2005)
Article CAS ADS PubMed Google Scholar - Cargill, M. et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nature Genet. 22, 231–238 (1999)
Article CAS PubMed Google Scholar - Ramensky, V., Bork, P. & Sunyaev, S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 30, 3894–3900 (2002)
Article CAS PubMed PubMed Central Google Scholar - Bittles, A. H. & Neel, J. V. The costs of human inbreeding and their implications for variations at the DNA level. Nature Genet. 8, 117–121 (1994)
Article CAS PubMed Google Scholar - Romeo, S. et al. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nature Genet. 39, 513–516 (2007)
Article CAS PubMed Google Scholar - International HapMap Consortium. A haplotype map of the human genome. Nature 437, 1299–1320 (2005)
- Voight, B. F. et al. Interrogating multiple aspects of variation in a full resequencing data set to infer human population size changes. Proc. Natl Acad. Sci. USA 102, 18508–18513 (2005)
Article CAS ADS PubMed PubMed Central Google Scholar - Ohta, T. Slightly deleterious mutant substitutions in evolution. Nature 246, 96–98 (1973)
Article CAS ADS PubMed Google Scholar - Travis, M. J. et al. Deleterious mutations can surf to high densities on the wave front of an expanding population. Mol. Biol. Evol. 24, 2334–2343 (2007)
Article CAS PubMed Google Scholar - Voight, B. F., Kudaravalli, S., Wen, X. & Pritchard, J. K. A map of recent positive selection in the human genome. PLoS Biol. 4, e72 (2006)
Article PubMed PubMed Central Google Scholar - Mekel-Bobrov, N. et al. Ongoing adaptive evolution of ASPM, a brain size determinant in Homo sapiens. Science 309, 1720–1722 (2005)
Article CAS ADS PubMed Google Scholar - Evans, P. D. et al. Microcephalin, a gene regulating brain size, continues to evolve adaptively in humans. Science 309, 1717–1720 (2005)
Article CAS ADS PubMed Google Scholar - Akey, J. M. et al. Population history and natural selection shape patterns of genetic variation in 132 genes. PLoS Biol. 2, e286 (2004)
Article PubMed PubMed Central Google Scholar - Slatkin, M. A population-genetic test of founder effects and implications for Ashkenazi Jewish diseases. Am. J. Hum. Genet. 75, 282–293 (2004)
Article CAS PubMed PubMed Central Google Scholar - Kere, J. Human population genetics: Lessons from Finland. Annu. Rev. Genomics Hum. Genet. 2, 103–128 (2001)
Article CAS PubMed Google Scholar - Kent, W. J., Baertsch, R., Hinrichs, A., Miller, W. & Haussler, D. Evolution’s cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl Acad. Sci. USA 100, 11484–11489 (2003)
Article CAS ADS PubMed PubMed Central Google Scholar - Karolchik, D. et al. The UCSC Genome Browser Database. Nucleic Acids Res. 31, 51–54 (2003)
Article CAS PubMed PubMed Central Google Scholar - Hernandez, R. D., Williamson, S. H. & Bustamante, C. D. Context dependence, ancestral misidentification, and spurious signatures of natural selection. Mol. Biol. Evol. 24, 1792–1800 (2007)
Article CAS PubMed Google Scholar - Kent, W. J. BLAT—the BLAST-like alignment tool. Genome Res. 12, 656–664 (2002)
Article CAS PubMed PubMed Central Google Scholar - Pruitt, K. D., Tatusova, T. & Maglott, D. R. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 33, D501–D504 (2005)
Article CAS PubMed Google Scholar - Wigginton, J. E., Cutler, D. J. & Abecasis, G. R. A note on exact tests of Hardy–Weinberg equilibrium. Am. J. Hum. Genet. 76, 887–893 (2005)
Article CAS PubMed PubMed Central Google Scholar - Hwang, D. G. & Green, P. Bayesian Markov chain Monte Carlo sequence analysis reveals varying neutral substitution patterns in mammalian evolution. Proc. Natl Acad. Sci. USA 101, 13994–14001 (2004)
Article CAS ADS PubMed PubMed Central Google Scholar
Acknowledgements
We thank the Celera Genomics sequencing centre, the International HapMap Consortium and SeattleSNPs for generation of these data sets. This work was supported by National Institutes of Health grants to A.G.C., C.D.B., R.N. and T. Matisse, and a National Science Foundation Graduate Research Fellowship to K.E.L.
Author Contributions K.E.L. and C.D.B. conceived of the original design of the project. J.J.S. and T.J.W. directed the collection of the sequence data by Celera Genomics. K.E.L., A.R.I., A.R.B., R.D.H., S.S. and M.J.H. designed the bioinformatics pipeline and analysed the data with direction from S.S., R.N., A.G.C. and C.D.B. K.E.L. performed the simulations. K.E.L., A.G.C. and C.D.B. wrote the paper with input from all authors.
Author information
Authors and Affiliations
- Department of Molecular Biology and Genetics,,
Kirk E. Lohmueller, Adam R. Boyko & Andrew G. Clark - Department of Biological Statistics and Computational Biology, Biotechnology Building, Cornell University, Ithaca, New York 14853, USA,
Kirk E. Lohmueller, Amit R. Indap, Adam R. Boyko, Ryan D. Hernandez & Carlos D. Bustamante - Department of Biochemistry, Max Planck Institute for Developmental Biology, 72076 Tübingen, Germany
Steffen Schmidt - Department of Human Genetics, University of Chicago, Chicago, Illinois 60637, USA,
Melissa J. Hubisz - Celera Diagnostics, Alameda, California 94592, USA,
John J. Sninsky & Thomas J. White - Division of Genetics, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts 02115, USA,
Shamil R. Sunyaev - Department of Biology, Center for Comparative Genomics, University of Copenhagen, Universitetsparken 15, 2100, KBH Ø, Denmark,
Rasmus Nielsen
Authors
- Kirk E. Lohmueller
You can also search for this author inPubMed Google Scholar - Amit R. Indap
You can also search for this author inPubMed Google Scholar - Steffen Schmidt
You can also search for this author inPubMed Google Scholar - Adam R. Boyko
You can also search for this author inPubMed Google Scholar - Ryan D. Hernandez
You can also search for this author inPubMed Google Scholar - Melissa J. Hubisz
You can also search for this author inPubMed Google Scholar - John J. Sninsky
You can also search for this author inPubMed Google Scholar - Thomas J. White
You can also search for this author inPubMed Google Scholar - Shamil R. Sunyaev
You can also search for this author inPubMed Google Scholar - Rasmus Nielsen
You can also search for this author inPubMed Google Scholar - Andrew G. Clark
You can also search for this author inPubMed Google Scholar - Carlos D. Bustamante
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toCarlos D. Bustamante.
Supplementary information
Supplementary Information
This file contains Supplementary Methods; Supplementary Figures 1-4; Supplementary Tables 1-5; Supplementary Notes 1-3 and additional references. (PDF 1020 kb)
Supplementary Data
This file contains Supplementary Data with the SNPs that were analyzed. The column headings are ‘ssid’, ‘hCV ID’, ‘Derived allele count AA’, ‘Total count AA’, ‘Derived allele count EA’, ‘Total count EA’, ‘functional prediction’. (TXT 1496 kb)
Rights and permissions
About this article
Cite this article
Lohmueller, K., Indap, A., Schmidt, S. et al. Proportionally more deleterious genetic variation in European than in African populations.Nature 451, 994–997 (2008). https://doi.org/10.1038/nature06611
- Received: 26 October 2007
- Accepted: 17 December 2007
- Issue Date: 21 February 2008
- DOI: https://doi.org/10.1038/nature06611