Sequencing the nuclear genome of the extinct woolly mammoth (original) (raw)
Main
Vertebrate genome sequencing projects have thus far assembled data from at least 28 species5, including chromosomal assemblies of six placental mammals, namely human6,7, chimpanzee8, rhesus macaque9, mouse10, rat11 and dog12. In contrast, kilobase (kb)-scale genomic sequence data from extinct species were first reported in 2005, with 27 kb from cave bear13 and 13 megabases (Mb) from mammoth14. More recently, two projects reported up to 1 Mb from Neanderthal15,16, some of which may be modern human contamination17.
Whereas many ancient-DNA studies have used bone samples, in 2007 we showed that DNA with fewer damage-induced substitutions can be extracted from hair shafts collected from permafrost remains18. Moreover, use of hair permits a highly efficient decontamination protocol that leaves the keratin-encased endogenous DNA unharmed. The method resulted in 15 complete mammoth mitochondrial genomes at high coverage18,19, identified in 947 Mb of total sequence (average read length, 93 bp) from a number of samples. Hair shafts are thus suitable for sequencing ancient nuclear DNA.
We selected M4, a Siberian mammoth specimen carbon-14 dated to 18,545 ± 70 years before present (roughly 20,000 years ago), for extensive sequencing, and generated 2.982 Gb of data from hair shafts using a Roche GS-FLX sequencing instrument. A second mammoth specimen, M25, yielded an additional 239 Mb. Together with our earlier mammoth data, this brought the total to 4.168 Gb of sequence. Given the abundance of hair available from M4 and M25, we were able to enrich the sequenced material for long DNA molecules, to overcome at least partly the high rate of breakage in ancient DNA. The average read length was 142 bp for M4 and 164 bp for M25. As a bonus, we obtained 4,430-fold coverage of the mitochondrial genome of M4, allowing us to determine error rates. (We assumed that errors in nuclear DNA equal those in mitochondrial DNA.) Specifically, for reads trimmed by aligning them to elephant sequence, the total error rate from post-mortem DNA damage and sequencing mistakes was 0.14%, neglecting errors that added or deleted bases (Table 1 and Methods).
Table 1 Basic statistics on the mammoth genome sequence
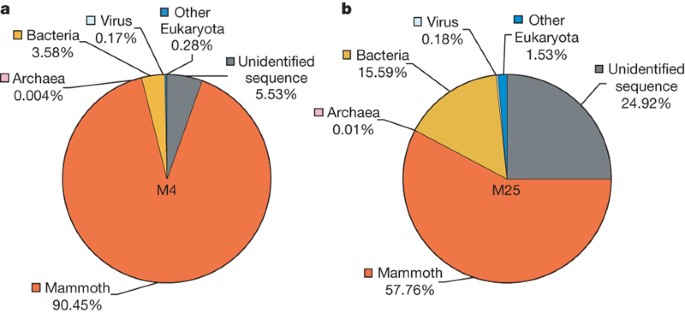
To estimate how many of our reads are actual mammoth DNA, we determined the fraction of our sequence that aligns to the African savanna elephant (Loxodonta africana) genome (twofold assembly and sixfold reads), which indicated that 80% of our 4.17 Gb of sequence, that is, approximately 3.3 Gb, is from the mammoth. However, the yield varied substantially between specimens: M4 is 90% mammoth and M25 is only 58% mammoth (Fig. 1). As a negative control, read-sized intervals of the chicken genome20 were mapped to elephant and showed that the false-positive rate is very low. Reasons why the estimation of 80% may actually be low are given in Methods. Some microbial DNA is recognizable in the non-mammoth portion (Fig. 1), but essentially none of the DNA in these samples is human18.
Figure 1: Species composition of metagenomic DNA extracted from mammoth hair.
a, b, Pie charts for the M4 (a) and M25 (b) data sets show the percentage of sequencing reads assigned to taxa for mammoth, Archaea, Bacteria, virus, as well as the two unspecified categories ‘other Eukaryota’ and ‘unidentified sequence’. The taxon distribution exemplifies the variability of the endogenous DNA content of ancient specimens.
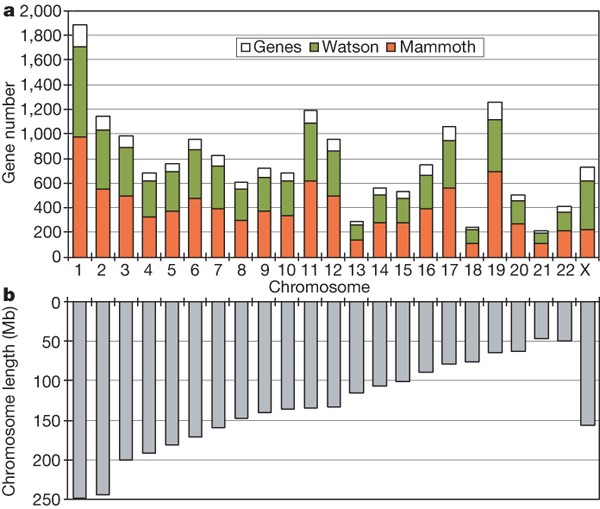
The converse result, that is, the fraction of elephant DNA that aligns to our data, can tell us how much of the mammoth genome has been sequenced. Because typical genome sizes of placental mammals are around 3 Gb, our 3.3 Gb might be expected to provide at least onefold coverage, in which case—taking into account overlapping reads21—over 63% of the bases in the mammoth genome would be sequenced at least once. However, the African elephant genome has previously been estimated at between 4.2 and 4.8 Gb using the C-value technique4, which, although less accurate than genome sequencing, has consistently predicted the Afrotherian genomes to be larger than previously sequenced placental genomes. We estimated how much of the mammoth genome has been sequenced by searching for matches to a set of elephant genes in the Ensembl gene build of August 2006 (http://www.ensembl.org) that could be confidently mapped to unique positions on human chromosomes (Fig. 2), and by searching for the so-called ultraconserved regions22. In both cases, around 50% of the bases were found; accounting for multiple reads that include the same genomic position, this translates into 0.7-fold coverage, or that the total length of our true mammoth reads is 70% of the genome’s length. Because some of our reads are very short and, hence, difficult to align reliably, this may be an underestimate.
Figure 2: Sequenced mammoth orthologues of human genes.
a, Plot showing the number of RefSeq genes on each human chromosome (open white rectangles), the average fraction of protein-coding bases that align to Roche/454 reads from James D. Watson’s genome30 (green), and the fraction of coding bases that align to one or more mammoth reads (red), using Ensembl-predicted elephant genes that map to the human chromosome—approximately 50% for each autosome, but only 31% for chromosome X as M4 was male (see Methods). b, Lengths of the chromosomes in a.
Our estimates that 80% of our 4.17 Gb of sequence is from mammoth and that we have obtained 0.7-fold coverage are consistent with a genome size of 4.7 Gb, as 4.17 × 0.8 ≈ 3.3 ≈ 4.7 × 0.7. However, this estimation of genome size is at best a rough approximation. On the other hand, we observed the probable cause of the expanded genome, namely an unusually high fraction of interspersed repeats (Supplementary Information).
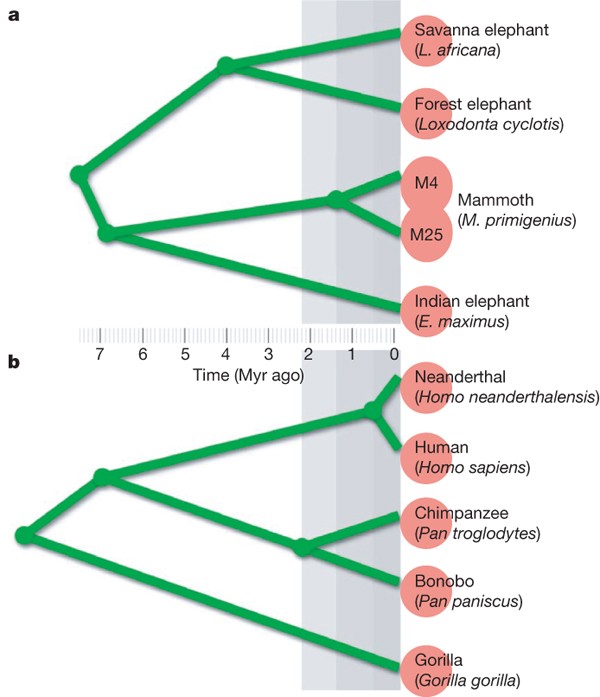
As currently understood, the evolutionary relationships among selected living and extinct elephantid species are sketched in Fig. 3; we show parallels with humans and some great apes to provide a widely familiar point of reference. Here we use estimated divergence times, which are times to the common ancestor averaged across the genome. This should be distinguished from population split times or, in the case of distinct species, speciation times. For instance, two modern European humans have a population split time of 0 yr but a mean divergence of at least 500,000 yr. This distinction is important for recent speciation events. For example, the mean divergence time between human and chimpanzee is at least 2 Myr longer than the speciation time23 (see Methods for details). We are interested in comparing sequence identity rates between elephantids and between apes.
Figure 3: Comparison of phylogenies.
a, Elephantids; b, hominoids. We show estimated divergence times, that is, times to the common ancestor averaged across autosomes (see Methods). Red circles at the leaves of the phylogenetic tree indicate discernable species. This distinction was not made for the two clades of mammoth (M4 and M25) based on the fossil record (merged red circles).
To estimate the level of nucleotide identity (ignoring gaps) between M4 and the African elephant, we analysed the large number of elephant positions that have more than one aligning mammoth read, to reduce the effect of errors in our sequence (Methods). The estimated identity is 99.4%. The 0.6% difference rate is approximately half of that estimated between human and chimpanzee (1.24%)24, despite the similarity in divergence times (Fig. 3). This indicates that nucleotide substitutions are fixed in recent elephantid lineages at only half of the rate in great apes and humans, mirroring an earlier observation about mitochondrial DNA25. Using a similar approach (Methods), we estimate that M4 and the African elephant are 99.78% identical at the amino-acid level.
Significantly, among the multiply sequenced differences between M4 and the African elephant, M25 agrees with the African elephant in 13.3% of the cases (that is, 327 of 2,451) in which we had a high-identity alignment to M25. Under the assumption that M4 and the African elephant differ by 15 Myr of evolution (7.5 Myr in each lineage), this corresponds to a separation of about 1.5–2.0 Myr between M4 and M25. We assume that only a small fraction of the differing positions are under selection. We note that a divergence of 1–2 Myr between M4 and M25 was estimated earlier19 on the basis of mitochondrial data, for which M25 agrees with the African elephant in 14.5% of the cases (114 of 792) where M4 disagrees with the African elephant. The concordance between nuclear and mitochondrial data is particularly noteworthy because population-genetic analysis of African elephants has shown that different relationships are inferred from mitochondrial sequence than from nuclear sequence26. M4 and M25 belong to differing clades of mammoths that were identified on the basis of short mitochondrial sequences19,27. However, morphological criteria distinguishing the two clades have not been established, similar to the case of two phenotypically identical groups of extant brown bears in Sweden that have differing mitotypes and share the same territory28.
A major reason for sequencing the woolly mammoth is to identify functionally important amino-acid differences between mammoth and elephant. It is unclear what fraction of amino-acid differences have functional consequences, but it is likely to be rather small; for instance, one estimation29 is that ∼20% of common human amino-acid variants are deleterious. To start looking for such differences, we combined computational criteria (designed to enrich for validity and functional importance) with PCR amplification and Sanger sequencing in M4, M25, African elephant and Indian elephant, Elephas maximus. Our initial screening yielded 92 putative differences (Fig. 4, Supplementary Information) that have also passed additional manual screening for undesirable attributes such as lack of conservation (notably homoplasy) at the critical mammoth position, potential confusion with paralogues, processed and unprocessed pseudogenes, and tandem or other duplicative debris. We found a number of cases in which mammoth differed from an amino acid that appeared to be otherwise invariant throughout placental evolution (Supplementary Information), which may suggest functional significance of the protein position and positive selection in the mammoth lineage.
Figure 4: Experimentally verified amino-acid differences among African elephant, Indian elephant, M4 and M25.
Non-synonymous coding single-nucleotide polymorphisms between African elephant and mammoth identified by computational mean, termed single-amino-acid polymorphisms (SAPs), were experimentally verified through PCR amplification and Sanger sequencing. Six SAP categories for splits specific to African elephant (AE), Indian elephant (IE), mammoth, M4 and M25 are shown, together with one category for heterozygosity and other polymorphisms. Gene names are for the putative human orthologue.
From the data set presented here, we conclude that a high-fidelity, high-coverage mammoth genome will be feasible once the genome sequence for the African elephant has been completed and 10–30-fold (depending on the sequencing technology) more mammoth sequence has been generated. From our data, we estimate that mammoth and elephant differ on average at about one residue per protein (roughly 20,000 positions proteome-wide) and that 90% of those differences are potentially identifiable by means of higher-coverage short-read sequencing alone (that is, without enriching sequenced material for coding DNA or Sanger resequencing; see Methods). Apart from comparing protein sequences, we hope to pinpoint DNA differences between mammoth and elephant in the non-repetitive genomic intervals, so it may even be possible to detect differences in gene-regulatory signals. The catalogue of differences, along with computational predictions of the differences most likely to have functional consequences, will provide a resource to facilitate direct observation of genetically orchestrated changes over evolutionary time, for example those associated with adaptation to cold environments, dietary changes and so on. In addition, the determination of an even larger number of synonymous changes in those protein-coding intervals will permit identification of genes and gene families under selection during mammoth evolution. As we have demonstrated here, these studies can be carried out on both clades of mammoth despite the specimens’ large difference in age. It will therefore become possible to conduct genome-wide studies on multiple individuals with the goal of understanding whether the observed coalescence time of 1.5–2.0 Myr between M4 and M25 in fact did generate two species of mammoth, or whether this process was terminated by premature extinction of the clade of M25.
Methods Summary
The 4.17 Gb of individual reads from our mammoth samples, along with the sequenced PCR products produced while studying SAPs, were placed in a freely Internet-accessible BLAST server (http://mammoth.psu.edu) that was used for some of the analyses described here, such as determining the fraction of putative elephant coding exons and mammalian ultraconserved intervals that align to a mammoth-sample read. In addition, sequence data from the African savanna elephant genome, produced by the Broad Institute, was a critical ingredient for our analysis. Early in the project, we used an assembly of the twofold-coverage data, downloaded from the University of California, Santa Cruz Genome Bioinformatics website (http://genome.ucsc.edu); later we employed individual Sanger reads that provide roughly sixfold coverage.
For whole-genome mammoth–elephant comparisons, we handled the problem of unmasked elephantid-specific interspersed repeats by aligning mammoth-sample reads to the twofold elephant scaffolds using the ‘dynamic masking’ feature of the BLASTZ alignment program (see Methods); only the reads that did not align in that preliminary step were aligned to the sixfold reads. Reads that aligned to a unique position in the twofold assembly, and specifically to a position itself aligned with high identity to a human RefSeq coding exon, were analysed to predict elephant/mammoth SAPs. We assumed that any given read is either all mammoth DNA or completely non-mammoth. One set of computational conditions, designed to enrich for substitutions of functional importance, was (1) that the putative amino-acid difference between mammoth and elephant occur in a run of five amino acids identical between human and elephant, and (2) that the substitution have a negative BLOSUM62 score.
Online Methods
Sample preparation
We employed the protocols described in a previous paper18, augmented as follows. M4 and M25 DNA were size-selected before DNA library construction by running the samples on a 2% unstained agarose gel along with a 100-bp DNA ladder (N3231S, New England Biolabs). The ladder was excised and stained for fragment visualization and the M4 and M25 DNA were excised between the 400-bp and 1,000-bp fragments that corresponded to the 100-bp DNA ladder. The samples were then purified using a QIAquick Gel Extraction Kit (QIAGEN) and used for library construction according to the manufacturer’s instructions (Roche Applied Sciences).
Error rate
We used the 4,430-fold coverage of the M4 mitochondrial genome (see Supplementary Fig. 1) to assess the sequencing error and post-mortem DNA damage to our sample, by comparing the individual reads with their consensus. We observed 72,951,869 matches, 103,181 mismatches, 51,587 erroneous missing bases and 97,661 erroneous extra bases (Supplementary Table 1). The total error rate was 0.345%. The most frequent damage-induced substitutions31,32 are C → T and G → A, with G → A most probably an artefact of the T4 polymerase enzyme in the DNA library construction33. Following an earlier paper31 we estimated the C → T DNA damage rate by subtracting the number of T → C transitions from the number of C → T transitions, and similarly for G → A. Together these account for 17.5% of the total errors, whereas other (putatively sequencing error) mismatches constitute 23.4%. The remaining 59.1% of the error consists of insertions/deletions in the reads, mostly in homopolymer runs. Thus, ignoring erroneous insertions and deletions, the fraction of incorrect bases is predicted to be 40.9% (that is, 17.5% + 23.4%) of 0.345%, or about 0.14%.
The fraction of reads from the mammoth genome
We randomly picked a set equivalent to a half-run on the Roche/454 instrument (roughly 30 Mb). The reads were aligned to the twofold elephant assembly using the BLASTZ program34 (scoring matches, 1; mismatches, -3; unaligned nucleotides, -1; local alignment of score, ≥30). Once a read was found to align to the elephant assembly, it was not compared to subsequent elephant sequences, to avoid aligning each interspersed or tandem repeat segment thousands of times. Reads that did not align to the twofold elephant assembly were compared with the sixfold elephant (Sanger) reads, requiring a gap-free alignment scoring of at least 30 when a match of 1 and a mismatch of -3. The reads that aligned to L. africana in one of these two steps, comprising 80.1% of the 4.17 Gb, were considered to be mammoth DNA. We also used this approach on just the M4 sample, just the M25 sample, and on read-sized intervals from the chicken genome20; the latter indicated that 1.2% of non-mammoth reads and 1.6% of their bases align to elephant under our criteria.
On the other hand, there are at least two reasons why the value of 80% may underestimate the amount of mammoth DNA. First, as about 1% of the human genome sequence is contained in a read-sized interval that is completely absent from chimpanzee, and vice versa8, 1% of the mammoth genome may be missed by our method because there is no orthologous L. africana sequence. Also, the Roche/454 technology that we used can sequence parts of the genome that are not represented in Sanger-based data such as that for elephant, for example an additional 1.4% of the human genome30.
Non-mammoth reads
The metagenomic results summarized in Fig. 1 (see Supplementary Information for details) were obtained using the MEGAN35 metagenome analysis software (http://www-ab.informatik.uni-tuebingen.de/software/megan/welcome.html).
The fraction of the genome that is sequenced
We downloaded 15,717 Ensembl elephant gene models (ftp.ensembl.org/pub/release-49/fasta/loxodonta_africana/cdna) and determined a subset of 4,131 that could be computationally assigned to a unique orthologous position in the human genome. These were compared with mammoth-sample reads using the mammoth BLAST server (http://mammoth.psu.edu), and alignments were filtered to require at least 97% identity. For autosomal genes, 51% of the elephant protein-coding bases were aligned to one or more mammoth reads by the nucleotide-based aligner blastn, where we required alignments to have at least 97% identity. On the other hand, only 30.9% of the coding bases from the X chromosome aligned (Fig. 2), reflecting the fact that M4 was male (see below); it is very likely that some of the other sequences were from females, which explains why the fraction for the X chromosome is a little more than half the autosomal fraction.
In another test, we aligned mammoth reads to the 481 so-called ultraconserved regions22—481 intervals (mostly not protein-coding) averaging 262 bp that are identical among human, mouse and rat and, hence, very likely to be observable in any other placental mammal. These intervals total 121,250 bp, and we aligned them using BLASTZ with the parameters T = 2, Y = 2,000, a scoring match of 100 and a mismatch of -200, and penalized a gap of length N by 400 + 100_N_. This time, we identified 47% of the bases in our data.
M4 was male
We identified a number of M4 reads that align to male-specific human genes36. For instance, we found a 286-bp read that aligns at 84% identity to the HSFY1 mRNA (GenBank accession no. NM_033108). As anticipated, neither the read nor the HSFY1 mRNA aligns to the current elephant assembly (sequenced from a female), even under much less stringent conditions.
Divergence times
For Fig. 3, the estimated divergence times within elephantids are25 as follows: African elephants diverged from mammoths and Asian elephants approximately 7.6 Myr ago, mammoths diverged from Indian elephants approximately 6.7 Myr ago, and African savannah elephants diverged from African forest elephants (Loxodonta cyclotis) approximately 4.0 Myr ago. The divergence time of the two mammoth clades, represented by M4 and M25, is estimated to be 1.5 Myr, on the basis of ref. 19 and data presented in this paper. For the human–Neanderthal divergence, we use a date of 400,000 years, concordant with the fossil record, and add 500,000 years for within-Neanderthal divergence, similar to that for modern humans. For the other hominoid divergence times, we use23,37 7.0 Myr for human and chimpanzee, 2.2 Myr for chimpanzee and bonobo (Pan paniscus) and 8.75 Myr for human and gorilla.
The nucleotide similarity between M4 and elephant
We limited consideration to M4 reads that aligned to a single position in the elephant assembly, and further required that the aligning portion be at least 100 bp and align with at least 97% identity. We looked for elephant positions (1) that aligned to more than one mammoth read, (2) that always aligned to the same nucleotide and (3) where neither the elephant nucleotide nor an aligning nucleotide is ambiguous (that is, ‘N’). (This approach discarded elephant positions that aligned to non-identical mammoth nucleotides; such positions accounted for 0.28% of the multiply aligned elephant positions, consistent with the prediction that if two mammoth nucleotides are aligned to a given elephant position, then one of them will be incorrect because of sequencing error or DNA damage (and hence the two will differ) about 0.28% of the time, as each one has a predicted substitution-error rate of 0.14%.) Finally, we required that the elephant position be aligned to mammoth reads that were generated on different days, to avoid the possibility of artefactual duplicates caused by our sample-preparation protocol. We found 45,039,470 such consistently aligning positions, of which 44,773,945 (99.41%) were identical between mammoth and elephant. To be more precise, this is the estimated rate of identity between mammoth and the current elephant assembly. Among the consistently aligning positions of M4 and African elephant, 2,451 were covered by M25 reads that aligned exactly once to elephant, with at least 97% identity over at least 100 bp; M5 agreed with elephant in 327 cases.
The amino-acid similarity between M4 and elephant
We identified 175,949 pairwise non-overlapping human RefSeq protein-coding exons (9,911,624 amino acids, not counting those split by an intron), where we required that the exon has the same reading frame in all annotated splice variants that contain it. Of these exons, 99,946 (4,284,551 amino acids) align without a gap or an improper stop codon to the twofold elephant assembly available in May 2007, using human–elephant alignments downloaded from the University of California, Santa Cruz Genome Bioinformatics browser38. Of these exons, 76,750 (containing 3,331,646 amino acids) had at least 85% amino-acid identity between human and elephant. To help eliminate matches between paralogues (but retaining orthologues), we worked with that reduced set.
Orthologous mammoth amino acids were predicted as follows. We limited consideration to 454-sequencer reads that aligned to a unique position of the elephant assembly, where we required at least 97% nucleotide identity (counting gaps) between elephant and the aligned portion of the read. Whenever a read overlapped an exon and, within the overlap, there were no gaps or ‘N’s in elephant or mammoth, we recorded the human-, elephant- and mammoth-aligned amino acids in the overlap (nucleotides before or after the reading frame were ignored). This procedure produced 1,537,885 amino-acid triples, including cases where a human–elephant position aligns to multiple mammoth reads. As with nucleotide differences (see above), we identified putative elephant amino-acid positions that appeared more than once among these triples, and were always aligned to the same predicted mammoth amino acid. There were 165,532 such cases, of which 165,164 (99.78%) were identical between mammoth and elephant.
Amino-acid differences
The Supplementary Information contains (1) our computational protocol for picking potentially interesting amino-acid differences between African elephant and M4, (2) our experimental methods for validating the predictions, (3) detailed information about the validated difference and (4) data about evolutionary conservation of the differences highlighted in Supplementary Table 5.
How many mammoth–elephant amino-acid differences can potentially be determined?
Human RefSeq genes contain just under 10 million amino acids (removing redundancy caused by splice isoforms), and hence we estimate the corresponding number for elephant to be 10 million. Assuming 99.8% amino-acid identity between elephant and mammoth, this gives roughly 20,000 amino-acid differences. To detect a mammoth–elephant difference using short reads (say onefold coverage with 200-bp reads and sixfold coverage with 40-bp reads), it is necessary that the read can be mapped to the elephant assembly with high confidence. This becomes difficult in portions of the assembly that align elsewhere in the assembly, say with at least 95% identity over at least 100 bp. We determined that roughly 9% of the human RefSeq coding intervals meet these conditions. On this basis, we anticipate that roughly 90% of our reads that intersect a protein-coding region can be reliably mapped to the completed elephant assembly.
Accession codes
Primary accessions
GenBank/EMBL/DDBJ
Data deposits
Sequence data are available at the NCBI database's trace archive section under accession number SRA001810.
References
- Höss, M., Pääbo, S. & Vereshchagin, N. K. Mammoth DNA sequences. Nature 370, 333 (1994)
Article Google Scholar - Hagelberg, E. et al. DNA from ancient mammoth bones. Nature 370, 333–334 (1994)
Article CAS Google Scholar - Pääbo, S. et al. Genetic analyses from ancient DNA. Annu. Rev. Genet. 38, 645–679 (2004)
Article Google Scholar - Redi, C. A. et al. Genome size: a novel genomic signature in support of Afrotheria. J. Mol. Evol. 64, 484–487 (2007)
Article CAS ADS Google Scholar - Miller, W. et al. 28-way vertebrate alignment and conservation track at the UCSC Genome Browser. Genome Res. 17, 1797–1808 (2007)
Article CAS Google Scholar - International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001)
- Venter, J. C. et al. The sequence of the human genome. Science 291, 1304–1351 (2001)
Article CAS ADS Google Scholar - The Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437, 69–87 (2005)
- Rhesus Macaque Genome Sequencing and Analysis Consortium. Evolutionary and biomedical insights from the rhesus macaque genome. Science 316, 222–234 (2007)
- Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002)
- Rat Genome Sequencing Project Consortium. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428, 493–541 (2004)
- Lindblad-Toh, K. et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438, 803–819 (2005)
Article CAS ADS Google Scholar - Noonan, J. P. et al. Genomic sequencing of Pleistocene cave bears. Science 309, 597–600 (2005)
Article CAS ADS Google Scholar - Poinar, H. N. et al. Metagenomics to palaeogenomics: large-scale sequencing of mammoth DNA. Science 311, 392–394 (2006)
Article CAS ADS Google Scholar - Noonan, J. P. et al. Sequencing and analysis of Neanderthal genomic DNA. Science 314, 1113–1118 (2006)
Article CAS ADS Google Scholar - Green, R. E. et al. Analysis of one million base pairs of Neanderthal DNA. Nature 444, 330–336 (2006)
Article CAS ADS Google Scholar - Wall, J. D. & Kim, S. K. Inconsistencies in Neanderthal genomic DNA sequences. PLoS Genet. 3, e175 (2007)
Article Google Scholar - Gilbert, M. T. P. et al. Whole-genome shotgun sequencing of mitochondria from ancient hair shafts. Science 317, 1927–1930 (2007)
Article CAS ADS Google Scholar - Gilbert, M. T. P. et al. Intraspecific phylogenetic analysis of Siberian woolly mammoths using complete mitochondrial genomes. Proc. Natl Acad. Sci. USA 105, 8327–8332 (2008)
Article CAS ADS Google Scholar - International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432, 695–716 (2004)
- Lander, E. S. & Waterman, M. S. Genomic mapping by fingerprinting random clones: A mathematical analysis. Genomics 2, 231–239 (1988)
Article CAS Google Scholar - Bejerano, G. et al. Ultraconserved elements in the human genome. Science 304, 1321–1325 (2004)
Article CAS ADS Google Scholar - Patterson, N. et al. Genetic evidence for complex speciation of humans and chimpanzees. Nature 441, 1103–1108 (2006)
Article CAS ADS Google Scholar - Chen, F. C. & Li, W. H. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet. 68, 444–456 (2001)
Article CAS Google Scholar - Rohland, N. et al. Proboscidean mitogenomics: chronology and mode of elephant evolution using mastodon as an outgroup. PLoS Biol. 5, e207 (2007)
Article Google Scholar - Roca, A. L. et al. Cytonuclear genomic dissociation in African elephant species. Nature Genet. 37, 96–100 (2005)
Article CAS Google Scholar - Barnes, I. et al. Genetic structure and extinction of the woolly mammoth, Mammuthus primigenius . Curr. Biol. 17, 1072–1075 (2007)
Article CAS Google Scholar - Taberlet, P. et al. Localization of a contact zone between two highly divergent mitochondrial DNA lineages of the brown bear Ursus arctus in Scandinavia. Conserv. Biol. 9, 1255–1261 (1995)
Article Google Scholar - Sunyaev, S. et al. Prediction of deleterious human alleles. Hum. Mol. Genet. 10, 591–597 (2001)
Article CAS Google Scholar - Wheeler, D. A. et al. The complete genome of an individual by massively parallel DNA sequencing. Nature 452, 872–876 (2008)
Article CAS ADS Google Scholar - Stiller, M. et al. Patterns of nucleotide misincorporations during enzymatic amplification and direct large-scale sequencing of ancient DNA. Proc. Natl Acad. Sci. USA 103, 13578–13584 (2006)
Article CAS ADS Google Scholar - Gilbert, M. T. P. et al. Recharacterization of ancient DNA miscoding lesions: Insights in the era of sequencing-by-synthesis. Nucleic Acids Res. 35, 1–10 (2007)
Article CAS Google Scholar - Brotherton, P. et al. Novel high-resolution characterization of ancient DNA reveals C > U-type base modification events as the sole cause of post mortem miscoding lesions. Nucleic Acids Res. 35, 5717–5728 (2007)
Article CAS Google Scholar - Schwartz, S. et al. Human-mouse alignments with Blastz. Genome Res. 13, 103–107 (2003)
Article CAS Google Scholar - Huson, D. H. et al. MEGAN analysis of metagenomic data. Genome Res. 17, 377–386 (2007)
Article CAS MathSciNet Google Scholar - Skaletsky, H. et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423, 825–837 (2003)
Article CAS ADS Google Scholar - Caswell, J. L. et al. Analysis of chimpanzee history based on genome sequence alignments. PLoS Genet. 4, e1000057 (2008)
Article Google Scholar - Kent, W. J. et al. The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002)
Article CAS Google Scholar
Acknowledgements
This sequencing-by-synthesis study was made possible through generous funding from Penn State University and Roche Applied Sciences. W.M. was supported by grant HG002238 from the National Human Genome Research Institute and S.C.S. is supported in part by the Gordon and Betty Moore Foundation. This project is funded in part under a grant from the Pennsylvania Department of Health using Tobacco Settlement Funds appropriated by the US legislature. The Pennsylvania Department of Health specifically disclaims responsibility for any analyses, interpretations or conclusions. We thank T. Gilbert for introducing us to the use of hair shafts as a source of ancient DNA, C. Grøndahl for providing Asian elephant hair samples, and M.Wilson for suggestions. This paper is dedicated to the memory of Andrei Sher.
Author information
Author notes
- Andrei Sher: Deceased
Authors and Affiliations
- Pennsylvania State University, Center for Comparative Genomics and Bioinformatics, 310 Wartik Building, University Park, Pennsylvania 16802, USA ,
Webb Miller, Daniela I. Drautz, Aakrosh Ratan, Barbara Pusey, Ji Qi, Arthur M. Lesk, Lynn P. Tomsho, Michael D. Packard, Fangqing Zhao & Stephan C. Schuster - Severtsov Institute of Ecology and Evolution, Russian Academy of Sciences, 33 Leninsky Prospect, 119071 Moscow, Russia ,
Andrei Sher - Zoological Institute, Russian Academy of Sciences, Universitetskaya Naberezhnaya 1, 199034 Saint Petersburg, Russia
Alexei Tikhonov - Center for Biomolecular Science and Engineering, University of California, Santa Cruz, California 95064, USA ,
Brian Raney - Broad Institute of MIT and Harvard, 7 Cambridge Center, Cambridge, Massachusetts 02142, USA ,
Nick Patterson, Kerstin Lindblad-Toh & Eric S. Lander - 454 Life Sciences, 20 Commercial Street, Branford, Connecticut 06405, USA ,
James R. Knight & Gerard P. Irzyk - Roche Diagnostics Corporation, 9115 Hague Road, Indianapolis, Indiana 46250-0414, USA ,
Karin M. Fredrikson, Timothy T. Harkins & Sharon Sheridan - Sperling Foundation, Eugene, Oregon 97405, USA ,
Tom Pringle
Authors
- Webb Miller
You can also search for this author inPubMed Google Scholar - Daniela I. Drautz
You can also search for this author inPubMed Google Scholar - Aakrosh Ratan
You can also search for this author inPubMed Google Scholar - Barbara Pusey
You can also search for this author inPubMed Google Scholar - Ji Qi
You can also search for this author inPubMed Google Scholar - Arthur M. Lesk
You can also search for this author inPubMed Google Scholar - Lynn P. Tomsho
You can also search for this author inPubMed Google Scholar - Michael D. Packard
You can also search for this author inPubMed Google Scholar - Fangqing Zhao
You can also search for this author inPubMed Google Scholar - Andrei Sher
You can also search for this author inPubMed Google Scholar - Alexei Tikhonov
You can also search for this author inPubMed Google Scholar - Brian Raney
You can also search for this author inPubMed Google Scholar - Nick Patterson
You can also search for this author inPubMed Google Scholar - Kerstin Lindblad-Toh
You can also search for this author inPubMed Google Scholar - Eric S. Lander
You can also search for this author inPubMed Google Scholar - James R. Knight
You can also search for this author inPubMed Google Scholar - Gerard P. Irzyk
You can also search for this author inPubMed Google Scholar - Karin M. Fredrikson
You can also search for this author inPubMed Google Scholar - Timothy T. Harkins
You can also search for this author inPubMed Google Scholar - Sharon Sheridan
You can also search for this author inPubMed Google Scholar - Tom Pringle
You can also search for this author inPubMed Google Scholar - Stephan C. Schuster
You can also search for this author inPubMed Google Scholar
Corresponding authors
Correspondence toWebb Miller or Stephan C. Schuster.
Supplementary information
Supplementary Information
This file contains Supplementary Figure SF1 with Legend, Supplementary Tables ST1-ST5 and Supplementary References (PDF 1001 kb)
PowerPoint slides
Rights and permissions
About this article
Cite this article
Miller, W., Drautz, D., Ratan, A. et al. Sequencing the nuclear genome of the extinct woolly mammoth.Nature 456, 387–390 (2008). https://doi.org/10.1038/nature07446
- Received: 12 May 2008
- Accepted: 22 September 2008
- Issue Date: 20 November 2008
- DOI: https://doi.org/10.1038/nature07446