The genomes of four tapeworm species reveal adaptations to parasitism (original) (raw)
Abstract
Tapeworms (Cestoda) cause neglected diseases that can be fatal and are difficult to treat, owing to inefficient drugs. Here we present an analysis of tapeworm genome sequences using the human-infective species Echinococcus multilocularis, E. granulosus, Taenia solium and the laboratory model Hymenolepis microstoma as examples. The 115- to 141-megabase genomes offer insights into the evolution of parasitism. Synteny is maintained with distantly related blood flukes but we find extreme losses of genes and pathways that are ubiquitous in other animals, including 34 homeobox families and several determinants of stem cell fate. Tapeworms have specialized detoxification pathways, metabolism that is finely tuned to rely on nutrients scavenged from their hosts, and species-specific expansions of non-canonical heat shock proteins and families of known antigens. We identify new potential drug targets, including some on which existing pharmaceuticals may act. The genomes provide a rich resource to underpin the development of urgently needed treatments and control.
Similar content being viewed by others
Main
Echinococcosis (hydatid disease) and cysticercosis, caused by the proliferation of larval tapeworms in vital organs1, are among the most severe parasitic diseases in humans and account for 2 of the 17 neglected tropical diseases prioritized by the World Health Organization[2](/articles/nature12031#ref-CR2 "World Health Organization. Neglected Tropical Diseases ( http://www.who.int/neglected_diseases/diseases/en/
) (2012)"). Larval tapeworms can persist asymptomatically in a human host for decades[3](/articles/nature12031#ref-CR3 "Eckert, J. & Deplazes, P. Biological, epidemiological, and clinical aspects of echinococcosis, a zoonosis of increasing concern. Clin. Microbiol. Rev. 17, 107–135 (2004)"), eventually causing a spectrum of debilitating pathologies and death[1](/articles/nature12031#ref-CR1 "Garcia, H. H., Moro, P. L. & Schantz, P. M. Zoonotic helminth infections of humans: echinococcosis, cysticercosis and fascioliasis. Curr. Opin. Infect. Dis. 20, 489–494 (2007)"). When diagnosed, the disease is often at an advanced stage at which surgery is no longer an option[4](/articles/nature12031#ref-CR4 "Brunetti, E., Kern, P. & Vuitton, D. A. Expert consensus for the diagnosis and treatment of cystic and alveolar echinococcosis in humans. Acta Trop. 114, 1–16 (2010)"). Tapeworm infections are highly prevalent worldwide[5](/articles/nature12031#ref-CR5 "Budke, C. M., White, A. C., Jr & Garcia, H. H. Zoonotic larval cestode infections: neglected, neglected tropical diseases? PLoS Negl. Trop. Dis. 3, e319 (2009)"), and their human disease burden has been estimated at 1 million disability-adjusted life years, comparable with African trypanosomiasis, river blindness and dengue fever. Furthermore, cystic echinococcosis in livestock causes an annual loss of US$2 billion[6](/articles/nature12031#ref-CR6 "Torgerson, P. R. & Macpherson, C. N. The socioeconomic burden of parasitic zoonoses: global trends. Vet. Parasitol. 182, 79–95 (2011)").Tapeworms (Platyhelminthes, Cestoda) are passively transmitted between hosts and parasitize virtually every vertebrate species7. Their morphological adaptations to parasitism include the absence of a gut, a head and light-sensing organs, and they possess a unique surface (tegument) that is able to withstand host-stomach acid and bile but is still penetrable enough to absorb nutrients7.
Tapeworms are the only one of three major groups of worms that parasitize humans, the others being flukes (Trematoda) and round worms (Nematoda), for which no genome sequence has been available so far. Here we present a high-quality reference tapeworm genome of a human-infective fox tapeworm Echinococcus multilocularis. We also present the genomes of three other species, for comparison; E. granulosus (dog tapeworm), Taenia solium (pork tapeworm), both of which infect humans, and Hymenolepis microstoma (a rodent tapeworm and laboratory model for the human parasite Hymenolepis nana). We have mined the genomes to provide a starting point for developing urgently needed therapeutic measures against tapeworms and other parasitic flatworms. Access to the complete genomes of several tapeworms will accelerate the pace at which new tools and treatments to combat tapeworm infections can be discovered.
The genomes and genes of tapeworms
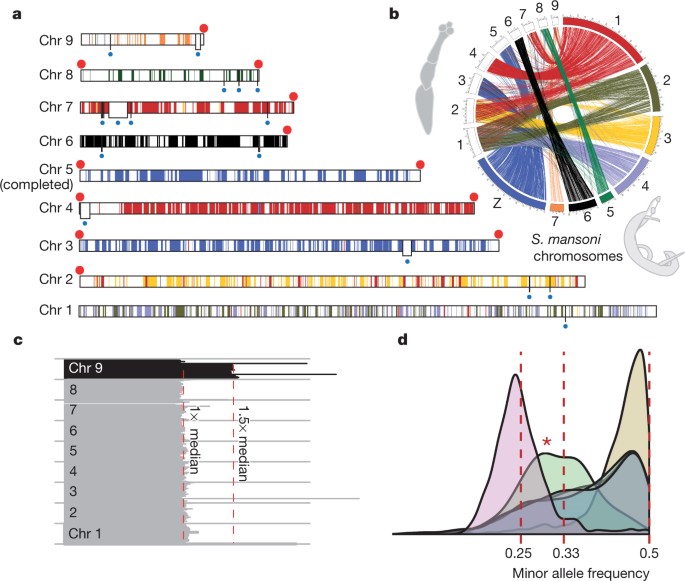
The E. multilocularis genome assembly was finished manually (Supplementary Information, section 2), producing a high-quality reference genome in which 89% of the sequence is contained in 9 chromosome scaffolds that have only 23 gaps (Supplementary Table 1.2). One chromosome is complete from telomere to telomere, and 13 of the expected 18 telomeres are joined to scaffolds (Fig. 1a). This quality and completeness is comparable to that of the first publications of Caenorhabditis elegans and Drosophila melanogaster genomes8,9. The 115- to 141-megabase (Mb) nuclear tapeworm genomes were sequenced using several high-throughput technologies (Supplementary Table 1.1). The tapeworm genomes are approximately one-third of the size of the genome of their distant flatworm relative, the blood fluke Schistosoma mansoni10, mainly because it has fewer repeats (Supplementary Information, section 3). By sequencing several isolates of E. multilocularis (Supplementary Table 3.2), we revealed tetraploidy in protoscoleces of one isolate, and a trisomy of chromosome 9 (the smallest chromosome, and possibly the only one for which a trisomy is tolerated) transiently exhibited in protoscoleces and metacestodes from two different isolates (Fig. 1c, d and Supplementary Figs 3.1, 3.2 and 3.3), consistent with previous observations of karyotype plasticity in flatworms11.
Figure 1: Genome of E. multilocularis.
a, The nine assembled chromosomes (Chr 1–Chr 9) of E. multilocularis with telomeres (red dots). Physical gaps in the sequence assembly (white boxes with blue dot beneath) are bridged by optical map data. Each colour segment is defined as an array of at least three genes that each has a single orthologous counterpart on one S. mansoni chromosome, regardless of their locations on the chromosome. b, One-to-one orthologues connecting E. multilocularis and S. mansoni chromosomes. c, Distribution of normalized genome coverage on isolate GT10/2. Each horizontal line depicts median coverage of 100-kb windows normalized against the mean coverage for the genome (130×). Even coverage was observed across the first eight chromosomes, but 1.5× coverage of chromosome 9 indicates trisomy. Similar plots for other isolates are shown in Supplementary Fig. 3.1. d, Distribution of minor allele frequency (MAF) of heterozygous sites in five isolates of E. multilocularis (plot for individual isolates in Supplementary Fig. 3.1), identified by mapping sequencing reads against the assembled chromosome consensus sequences. At each site, the proportion of bases that disagree with the reference is counted. For four isolates, the MAF peaks at around 0.5, indicative of diploidy, whereas JAVA05/1 peaks at 0.25, suggesting tetraploidy. Chromosome 9 of GT10/2 is plotted separately (marked by asterisk) from chromosomes 1 to 8, and the MAF display a clear departure of 0.5 and peaks around 0.33, consistent with a trisomy.
Aided by deep transcriptome sequencing from multiple life-cycle stages, we identified 10,231 to 12,490 putative genes per genome (Supplementary Table 5.5). Similar to the genome of S. mansoni12, distinct ‘micro-exon genes’ are present in tapeworm genomes, with multiple internal exons that are small (typically less than 36 bases) and divisible by 3 (Supplementary Information, section 5). To identify gene gain and loss in tapeworms, orthologous relationships were predicted between tapeworms and eight other species (Fig. 2). Although gene order has been lost, ancient chromosomal synteny is preserved among parasitic flatworms (Fig. 1b and Supplementary Table 7.3). Two chromosomes in E. multilocularis (Fig. 1a, b) correspond to the S. mansoni Z sex chromosome. Schistosomes are unusual among flatworms in that they have sexual dimorphism, but how common ancestors of both tapeworms and flukes evolved into female-heterogametic parasites, like S. mansoni, remains to be elucidated.
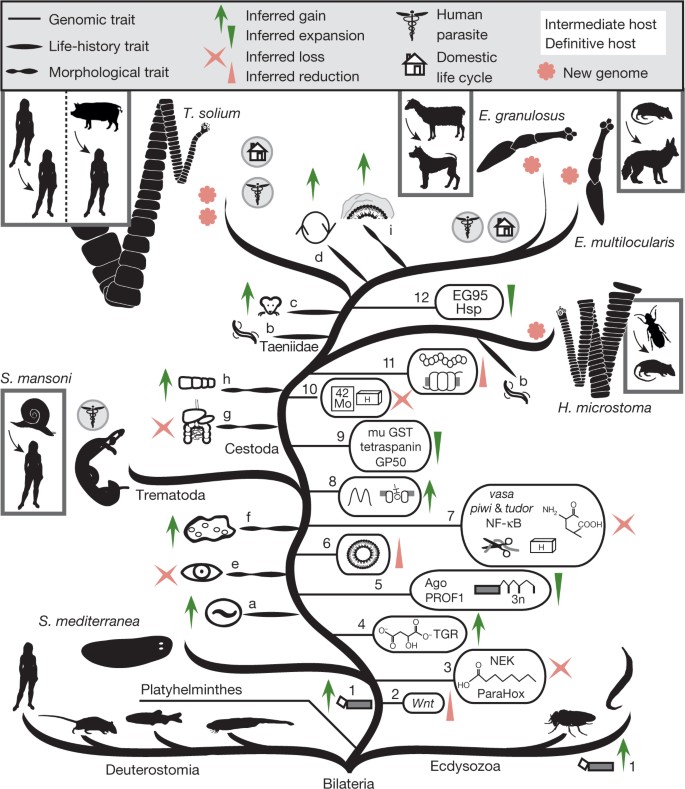
Figure 2: Evolution of tapeworm parasitism.
Phylogeny of the main branches of Bilateria; Ecdysozoa (including fruitflies and nematodes), Deuterostomia (including lancelet, zebrafish, mice and humans), and Lophotrochozoans (including Platyhelminthes (flatworms)) (based on phylogeny in Supplementary Fig. 7.1). The gains and losses of life-cycle traits for these parasitic flatworms include the evolution of endoparasitism (a), passive transmission between hosts (b), acquisition of vertebrate intermediate host (c), ability to proliferate asexually in intermediate host (d). Morphological traits that have evolved include the loss of eye cups (e), gain of neodermatan syncytial epithelia (f), loss of gut (g), segmentation of body plan (h), and changes in the laminated layer (to contain specialized apomucins; i). Gains and losses of genomic traits include spliced-leader _trans_-splicing (1), loss of Wnt genes (2), loss of NEK kinases, fatty acid biosynthesis and ParaHox genes (3), anaerobic metabolic ability through the malate dismutation/rodhoquinone pathway, merger of glutaredoxin and thioredoxin reductase to thioredoxin glutathione reductase (TGR) (4), evolution of tapeworm- and fluke-specific Argonaute (Ago) family, micro exon genes (MEGs) and PROF1 GPCRs (5), loss of peroxisomal genes (6), and complete loss of vasa, tudor and piwi genes, NF-κB pathway, loss of 24 homeobox gene families (indicated by ‘H’), metabolic proteases and amino acid biosynthesis (7). In tapeworms, gains and losses of genomic traits include innovation of bimodal intron distribution and novel fatty acid transporters (8), expansion of mu-class glutathione _S_-transferases, GP50 antigens and tetraspanins (9), loss of the molybdopterin biosynthesis pathway, loss of 10 homeobox gene families (10), fewer GPCRs and fewer neuropeptides encoded by each protopeptide (11), and expansion of heat shock proteins (Hsp) and species-specific antigens (12).
Genome-wide identification of polycistrons in tapeworms shows that there are 308 putative polycistrons in E. multilocularis, with the largest containing 4 genes. The internal gene order within E. multilocularis polycistrons is largely the same as in T. solium and H. microstoma (Supplementary Table 6.5), and—to some extent—as in flukes; 39% of S. mansoni orthologues of genes within E. multilocularis polycistrons retain colinearity. Of these S. mansoni genes, 40% have transcriptome evidence supporting their polycistronic transcription10, demonstrating further that gene order in polycistrons is highly conserved over long evolutionary time13 (P < 0.0001, Supplementary Information, section 6).
Polycistrons are resolved into individual coding transcripts using spliced-leader _trans_-splicing, but spliced-leader _trans_-splicing also occurs in genes outside of polycistrons. Using deep transcriptome sequencing (RNA-seq) we found evidence of spliced-leader _trans_-splicing in approximately 13% of E. multilocularis genes (Supplementary Table 6.2), less than the 70% observed in C. elegans14 and 58% in a tunicate15.
Specialized metabolism and detoxification
The high-confidence gene sets reveal extensive reductions in overall metabolic capability and an increased ability to absorb nutrients, compared to that of other animals (Figs 2 and 3, and Supplementary Information, section 9). Their main energy source, carbohydrates, can be catabolized by aerobic respiration or by two complementary anaerobic pathways; the lactate fermentation and malate dismutation pathways. The parasiticidal effects of mitochondrial fumarate reductase inhibitors have been demonstrated in vitro, suggesting that the malate dismutation pathway would be an effective target for the development of novel therapeutics16.
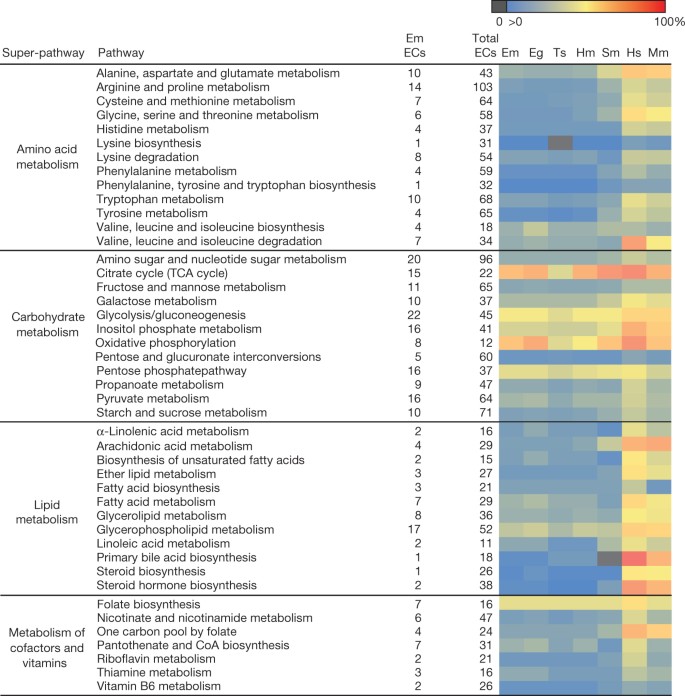
Figure 3: Conservation of individual metabolic pathways.
Heatmap showing the conservation of individual metabolic pathways for E. multilocularis (Em), E. granulosus (Eg), T. solium (Ts), H. microstoma (Hm) and S. mansoni (Sm) compared to those of humans (Hs) and mice (Mm). Each row indicates an individual metabolic pathway grouped by their superclass membership (defined by KEGG (Kyoto Encyclopedia of Genes and Genomes)). Coloured tiles indicate the level of conservation (percentage of enzymes detected) of each pathway within each species. KEGG pathways with insufficient evidence (that is, containing only one enzyme) in E. multilocularis have been removed. CoA, coenzyme A; EC, enzyme commission number; TCA, tricarboxylic acid cycle.
Tapeworms, like flukes, lack the ability to synthesize fatty acids and cholesterol de novo17,18. Instead, they scavenge essential fats from the host using fatty acid transporters and lipid elongation enzymes (Supplementary Table 9.2), as well as several tapeworm-specific gene families, such as fatty acid binding protein (FABP) and the apolipoprotein antigen B (Supplementary Information, section 8). Uptake of fatty acids seems to be crucial in Echinococcus spp. metacestodes, in which both FABP and antigen B gene families are among the most highly expressed genes19 (Supplementary Table 5.7). Tapeworms and flukes have lost many genes associated with the peroxisome (Supplementary Information, section 8), an organelle in which fatty acid oxidation occurs, and may lack peroxisomes altogether, as seen in several other parasites20.
Compared with other animals, S. mansoni has a reduced ability to synthesize amino acids17. In tapeworms, this capacity is reduced further, with serine and proline biosynthesis enzymes absent from E. multilocularis (Fig. 3 and Supplementary Information, section 9). Many enzymes in the molybdopterin biosynthesis pathway seemed to be lost in tapeworms, along with enzymes that use molybdopterin as a cofactor. The ability to utilize molybdenum in enzymatic reactions was believed to be present in all animals21, but has been lost in some eukaryotic parasites22.
Differences in the detoxification systems between tapeworms and their mammalian hosts may be exploited for drug design (Supplementary Information, section 9). We found that, like flukes23, tapeworms typically have only one cytochrome P450 gene, suggesting that their ability to oxidize many xenobiotics and steroids is substantially lower than that of their hosts. Uniquely, tapeworms and flukes have merged two key enzymatic functions for redox homeostasis in one single enzyme: thioredoxin glutathione reductase (TGR). TGR is an essential gene and validated drug target in flukes24. Downstream of TGR we find an unexpected diversity of thioredoxins, glutaredoxins and mu-class glutathione S-transferases (GSTs) (Supplementary Table 9.3). The GST expansion suggests that tapeworms would be able to water-solubilize and excrete a large range of hydrophobic compounds, which may add complexity to the pharmacokinetics of drugs.
Homeobox gene loss
Homeobox genes are high-level transcription factors that are implicated in the patterning of body plans in animals. Across parasitic flatworms, the homeobox gene numbers are extensively reduced (Supplementary Table 10.1). Most bilaterian invertebrates have a conserved set of approximately 100 homeobox genes (for example, 92 conserved in C. elegans, 102 in D. melanogaster, and 133 in the lancelet)25. Of the 96 homeobox gene families that are thought to have existed at the origin of the Bilateria, 24 are not present in tapeworms and flukes, and a further 10 were lost in tapeworms, making their complement by far the most reduced of any studied bilaterian animal25. Among the tapeworm-specific gene losses are gene families involved in neural development (mnx, pax3/7, gbx, hbn and rax). This is somewhat surprising considering that tapeworms possess a well-developed nervous system, albeit with reduced sensory input and cephalization. Tapeworms also lack the ParaHox genes (gsx, pdx, cdx) ancestrally involved in specification of a through-gut26,27, although these seem to have been lost before the tapeworm gut was lost. Other conserved genes found in bilaterian developmental pathways such as Hedgehog and Notch were found to be present and intact, although the Wnt complement is greatly reduced compared to the ancestral (spiralian) complement of 12 Wnt ligands28 (Supplementary Table 10.2).
Stem cell specializations
Extreme regenerative capability and developmental plasticity, mediated by ever-present somatic stem cells (neoblasts), have made flatworms popular models for stem cell research29. All multicellular organisms rely on stem cells for proliferation and growth, so it is remarkable that tapeworms and flukes appear to lack the ubiquitous stem cell marker gene vasa (Supplementary Information, section 11). Instead tapeworms have two copies of another dead-box helicase (PL10), which we propose may have taken over some of the functions of vasa (Supplementary Fig. 11.1). Tapeworms and flukes are also missing the piwi gene subfamily and _piwi_-interacting _tudor_-domain containing proteins. The piwi genes belong to a subfamily of genes encoding argonaute proteins, and we also found that tapeworms have a new subfamily of argonaute proteins (Supplementary Fig. 11.2) that may bind a newly discovered potential small RNA precursor30. Both piwi and vasa are usually essential in regulating the fate of germline stem cells in animals, and vasa suppression usually leads to infertility or death31. These findings suggest that stem-cell-associated pathways in parasitic flatworms may be highly modified.
Specialization of the tapeworm proteome
We sought to identify novel and expanded gene families in tapeworms, and found many frequently occurring novel domains involved in cell–cell adhesion and the formation of the tegument (Supplementary Information, section 8). For example, several novel domains are found on the ectodomain of cadherins (Supplementary Information, section 8), and tapeworms have proportionally more tetraspanin copies (30–36) (Supplementary Table 12.1) than the highly expanded repertoires of fruitflies and zebrafish32. The acellular carbohydrate-rich laminated layer, which coats the outside of Echinococcus metacestodes, is a unique genus-specific trait and one of the few morphological traits that differ between the very closely related species E. granulosus and E. multilocularis. We identified corresponding species differences in an _Echinococcus_-specific apomucin family (Supplementary Fig. 12.1), an important building block of the laminated layer33. One particular copy is highly differentiated between the two species (non-synonymous to synonymous substitution ratio of >1) and is the fifth most highly expressed in the metacestode stage of E. multilocularis (Supplementary Table 5.7). Galactosyltransferases that probably decorate the apomucins with galactose residues, the predominant sugar of laminated layer glycans, are similarly diverged33 (Supplementary Information, section 8). Approximately 20% of the genes are exclusive to tapeworms, and these include many highly expressed antigen families, such as antigen B, the glycosylphosphatidylinositol (GPI)-anchored protein GP50 (ref. 34), and the vaccine target EG95 (ref. 35) (Supplementary Table 12.4).
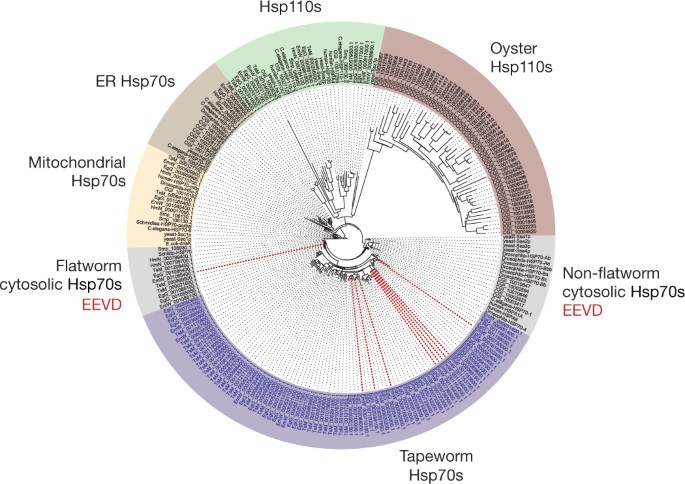
One of the most striking gene family expansions in the tapeworm genomes is the heat shock protein 70 (Hsp70) family. Phylogenetic analysis revealed independent and parallel expansions in both the Hsp110 and the cytosolic Hsp70 clades (Fig. 4). Several examples of expansions exist at various clades of Hsp70 in other systems, including Hsp110 expansions in oysters (to cope with temperature) and in cancer cells (to cope with proteotoxic stress)36,37. Echinococcus and T. solium have the highest number of gene expansions in the cytosolic Hsp70 clade. These expansions seem to have occurred independently in each species, and have resulted in 22 to 32 full copies in each species (Echinococcus and T. solium) compared to 6 copies in fruitflies and 2 in humans (Fig. 4). This expanded clade lacks classical cytosolic Hsp70 features (a conserved EEVD motif for substrate binding and a GGMP repeat unit), and whereas the canonical cytosolic hsp70 genes are constitutively expressed in different life-cycle stages, the non-canonical genes show almost no expression, suggesting a putative contingency role in which individual copies of the expanded family are only highly expressed under certain conditions (Supplementary Fig. 12.2). At least 40% of E. multilocularis _hsp70_-like genes are found within the subtelomeric regions of chromosomes, including the extreme case of chromosome 8 in which eight copies (including pseudogenes) are located in the subtelomere (Supplementary Table 12.2). No other genes are over-represented in these regions. Although Hsp70 proteins have been found in excretory–secretory products of tapeworms38, it remains to be determined whether the non-canonical Hsps have a host-interacting role or whether telomere proximity is important for their function or expression.
Figure 4: Heat shock protein 70 expansions in tapeworms.
Rooted tree of Hsp70 sequences from tapeworms and the eight comparator species used in this study, with additional sequences from baker’s yeast Saccharomyces cerevisiae, and the Pacific oyster Crassostrea gigas (a non-flatworm example of a lophotrochozoan with a recently reported Hsp70 expansion). Different Hsp70 subfamilies are shown in different colours. Dotted red lines, E. multilocularis hsp70 genes that are located in the subtelomeres. EEVD, the conserved carboxy-terminal residues of a canonical cytosolic Hsp70; ER Hsp70, endoplasmic reticulum Hsp70.
Novel drug targets
Tapeworm cysts are treated by chemotherapy or surgical intervention depending on tapeworm species, patient health and the site of the cyst. The only widely used drugs to treat tapeworm cysts are benzimidazoles39 that, owing to considerable side effects, are administered at parasitistatic rather than parasiticidal concentrations40. Novel targets and compound classes are therefore urgently needed.
To identify new potential drug targets, we surveyed common targets of existing pharmaceuticals; kinases, proteases, G-protein-coupled receptors (GPCRs) and ion channels41. We identified approximately 250 to 300 new protein kinases (Supplementary Table 13.1), and these cover most major classes (Supplementary Information, section 13). We also identified 151 proteases and 63 peptidase-like proteins in E. multilocularis, a repertoire of similar diversity to S. mansoni, and found that, like S. mansoni, E. multilocularis has strongly reduced copy numbers compared to those of other animals (Supplementary Table 13.9). Many successful anthelminthic drugs target one of several different forms of neural communication41. We therefore mapped the signalling pathways of the serotonin and acetylcholine neurotransmitters, predicted conserved and novel neuropeptides (Supplementary Table 13.6), and classified more than 60 putative GPCRs (Supplementary Table 13.2) and 31 ligand-gated ion channels (Supplementary Table 13.4). A voltage-gated calcium channel subunit42—the proposed target of praziquantel—is not expressed in cysts and thus provides a putative explanation for the drug’s low efficacy.
We searched databases for potential features for target selection, including compounds associated with protein targets and expression in the clinically relevant metacestode life-stage, and using this information we assigned weights to rank the entire proteomes (Supplementary Table 13.10). We identified 1,082 E. multilocularis proteins as potential targets, and of these, 150 to 200 with the highest scores have available chemical leads (known drug or approved compounds).
Acetylcholinesterases, which are inhibited by mefloquine (an anti-malarial that reduces egg production in S. mansoni), are high on the list of potential targets43. However, acetylcholinesterase transcription in tapeworm cysts is low, possibly limiting their suitability. After filtering to remove targets with common substrates rather than inhibitors, the top of the list includes several homologues of targets for cancer chemotherapy, including casein kinase II, ribonucleoside reductase, UMP–CMP kinase and proteasome subunits (Table 1). The challenges of inhibiting cancer tumours and metacestodes (particularly those of E. multilocularis) with drugs are in some ways similar; both show uncontrolled proliferation, invasion and metastasis, and are difficult to kill without causing damage to the surrounding tissue. Therefore, metacestodes may be vulnerable to similar strategies as cancer; suppression of mitosis, induction of apoptosis and prevention of DNA replication. In fact, the anthelminthic medicines niclosamide, mebendazole and albendazole have already been shown to inhibit cancer growth44.
Table 1 Top 20 promising targets in E. multilocularis
Conclusion
Tapeworms were among the first known parasites of humans, recorded by Hippocrates and Aristotle in ∼300 bc (ref. 45), but a safe and efficient cure to larval tapeworm infection in humans has yet to be found. These genomes provide hundreds of potential drug targets that can be tested using high-throughput drug screenings that were made possible by recent advances in axenic and cell culturing techniques39,46,47. Flatworms display an unusually high degree of developmental plasticity. In this study, the high level of sequence completion enabled both gene losses and gains to be accurately determined, and has shown how this plasticity has been put to use in the evolution of tapeworms.
Methods Summary
Genome sequencing was carried out using a combination of platforms. RNA sequencing was performed with Illumina RNA-seq protocols (for E. multilocularis, E. granulosus and H. microstoma) or capillary sequencing of full-length complementary DNA libraries (T. solium). The complete genome annotation is available at http://www.genedb.org. The tapeworm genome projects were registered under the INSDC project IDs PRJEB122 (E. multilocularis), PRJEB121 (E. granulosus), PRJEB124 (H. microstoma) and PRJNA16816 (T. solium). Sequence data for T. solium isolate (from Mexico) were used for all orthologue comparisons, but results relating to gene gains and losses were reconciled against an additional sequenced isolate from China (unpublished). All experiments involving jirds (laboratory host of E. multilocularis) were carried out in accordance with European and German regulations relating to the protection of animals. Ethical approval of the study was obtained from the ethics committee of the government of Lower Franconia (621-2531.01-2/05). Experiments with dogs (host of E. multilocularis sample RNA-seq ERS018054) were conducted according to the Swiss guidelines for animal experimentation and approved by the Cantonal Veterinary Office of Zurich prior to the start of the study, and were carried out with facility-born animals at the experimental units of the Vetsuisse Faculty in Zurich (permission numbers 40/2009 and 03/2010). A licensed hunter hunted the fox (host of E. multilocularis sample RNA-seq ERS018053) during the regular hunting season. Hymenolepis parasites were reared using laboratory mice in accordance with project license PPL 70/7150, granted to P.D.O. by the UK Home Office.
Accession codes
Primary accessions
ArrayExpress
European Nucleotide Archive
Expressed Sequence Tag Database
Data deposits
The tapewormgenome projects were registered under the INSDC project IDs PRJEB122 (E. multilocularis), PRJEB121 (E. granulosus), PRJEB124 (H. microstoma) and PRJNA16816 (T. solium, Mexico). Illumina and 454 data are released to the European Nucleotide Archive (http://www.ebi.ac.uk/ena/) under accession numbers ERP000351, ERP000452 and PRJNA16816. Capillary data is at http://www.ncbi.nlm.nih.gov/Traces/trace.cgi, SEQ_LIB_ID 98488, 98489 and 101760, CENTER NAME SC (E. multilocularis) and sg1, sg2, sg3, sg4 and sg5 (T. solium, Mexico). Genome data are available from http://www.sanger.ac.uk/resources/downloads/helminths/ (E. multilocularis, E. granulosus and H. microstoma) and http://www.taeniasolium.unam.mx/taenia/ (T. solium). The complete genome annotation is available at http://www.genedb.org. All RNA-seq data were released to ArrayExpress under accession numbers E-ERAD-50 or E-ERAD-56. T. solium EST sequences were released to http://www.ncbi.nlm.nih.gov/nucest/, under accession numbers EL740221 to EL763490.
References
- Garcia, H. H., Moro, P. L. & Schantz, P. M. Zoonotic helminth infections of humans: echinococcosis, cysticercosis and fascioliasis. Curr. Opin. Infect. Dis. 20, 489–494 (2007)
Article Google Scholar - World Health Organization. Neglected Tropical Diseases (http://www.who.int/neglected_diseases/diseases/en/) (2012)
- Eckert, J. & Deplazes, P. Biological, epidemiological, and clinical aspects of echinococcosis, a zoonosis of increasing concern. Clin. Microbiol. Rev. 17, 107–135 (2004)
Article Google Scholar - Brunetti, E., Kern, P. & Vuitton, D. A. Expert consensus for the diagnosis and treatment of cystic and alveolar echinococcosis in humans. Acta Trop. 114, 1–16 (2010)
Article Google Scholar - Budke, C. M., White, A. C., Jr & Garcia, H. H. Zoonotic larval cestode infections: neglected, neglected tropical diseases? PLoS Negl. Trop. Dis. 3, e319 (2009)
Article Google Scholar - Torgerson, P. R. & Macpherson, C. N. The socioeconomic burden of parasitic zoonoses: global trends. Vet. Parasitol. 182, 79–95 (2011)
Article Google Scholar - Burton, J., Bogitsh, C. E. C. & Oeltmann, T. N. Human parasitology. 4th edn (Academic Press, 2012)
Google Scholar - Adams, M. D. et al. The genome sequence of Drosophila melanogaster. Science 287, 2185–2195 (2000)
Article Google Scholar - The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012–2018 (1998)
- Protasio, A. V. et al. A systematically improved high quality genome and transcriptome of the human blood fluke Schistosoma mansoni. PLoS Negl. Trop. Dis. 6, e1455 (2012)
Article CAS Google Scholar - Špakulová, M., Orosova, M. & Mackiewicz, J. S. Cytogenetics and chromosomes of tapeworms (Platyhelminthes, Cestoda). Adv. Parasitol. 74, 177–230 (2011)
Article Google Scholar - DeMarco, R. et al. Protein variation in blood-dwelling schistosome worms generated by differential splicing of micro-exon gene transcripts. Genome Res. 20, 1112–1121 (2010)
Article CAS Google Scholar - Qian, W. & Zhang, J. Evolutionary dynamics of nematode operons: easy come, slow go. Genome Res. 18, 412–421 (2008)
Article CAS Google Scholar - Allen, M. A., Hillier, L. W., Waterston, R. H. & Blumenthal, T. A global analysis of C. elegans trans-splicing. Genome Res. 21, 255–264 (2011)
Article CAS Google Scholar - Matsumoto, J. et al. High-throughput sequence analysis of Ciona intestinalis SL trans-spliced mRNAs: alternative expression modes and gene function correlates. Genome Res. 20, 636–645 (2010)
Article CAS Google Scholar - Matsumoto, J. et al. Anaerobic NADH-fumarate reductase system is predominant in the respiratory chain of Echinococcus multilocularis, providing a novel target for the chemotherapy of alveolar echinococcosis. Antimicrob. Agents Chemother. 52, 164–170 (2008)
Article CAS Google Scholar - Berriman, M. et al. The genome of the blood fluke Schistosoma mansoni. Nature 460, 352–358 (2009)
Article ADS CAS Google Scholar - Frayha, G. J. Comparative metabolism of acetate in the taeniid tapeworms Echinococcus granulosus, E. multilocularis and Taenia hydatigena. Comp. Biochem. Physiol. B 39, 167–170 (1971)
Article CAS Google Scholar - Obal, G. et al. Characterisation of the native lipid moiety of Echinococcus granulosus antigen B. PLoS Negl. Trop. Dis. 6, e1642 (2012)
Article CAS Google Scholar - Kaasch, A. J. & Joiner, K. A. Targeting and subcellular localization of Toxoplasma gondii catalase. Identification of peroxisomes in an apicomplexan parasite. J. Biol. Chem. 275, 1112–1118 (2000)
Article CAS Google Scholar - Schwarz, G. & Mendel, R. R. Molybdenum cofactor biosynthesis and molybdenum enzymes. Annu. Rev. Plant Biol. 57, 623–647 (2006)
Article CAS Google Scholar - Zhang, Y., Rump, S. & Gladyshev, V. N. Comparative Genomics and Evolution of Molybdenum Utilization. Coord. Chem. Rev. 255, 1206–1217 (2011)
Article CAS Google Scholar - Pakharukova, M. Y. et al. Cytochrome P450 in fluke Opisthorchis felineus: identification and characterization. Mol. Biochem. Parasitol. 181, 190–194 (2012)
Article CAS Google Scholar - Kuntz, A. N. et al. Thioredoxin glutathione reductase from Schistosoma mansoni: an essential parasite enzyme and a key drug target. PLoS Med. 4, e206 (2007)
Article Google Scholar - Zhong, Y. F. & Holland, P. W. HomeoDB2: functional expansion of a comparative homeobox gene database for evolutionary developmental biology. Evol. Dev. 13, 567–568 (2011)
Article Google Scholar - Holland, P. W. Beyond the Hox: how widespread is homeobox gene clustering? J. Anat. 199, 13–23 (2001)
Article CAS Google Scholar - Brooke, N. M., Garcia-Fernandez, J. & Holland, P. W. The ParaHox gene cluster is an evolutionary sister of the Hox gene cluster. Nature 392, 920–922 (1998)
Article ADS CAS Google Scholar - Riddiford, N. & Olson, P. D. Wnt gene loss in flatworms. Dev. Genes Evol. 221, 187–197 (2011)
Article CAS Google Scholar - Brehm, K. Echinococcus multilocularis as an experimental model in stem cell research and molecular host-parasite interaction. Parasitology 137, 537–555 (2010)
Article CAS Google Scholar - Parkinson, J. et al. A transcriptomic analysis of Echinococcus granulosus larval stages: implications for parasite biology and host adaptation. PLoS Negl. Trop. Dis. 6, e1897 (2012)
Article Google Scholar - Raz, E. The function and regulation of vasa-like genes in germ-cell development. Genome Biol. 1, R1017.1–R1017.6 (2000)
Article Google Scholar - Garcia-España, A. et al. Appearance of new tetraspanin genes during vertebrate evolution. Genomics 91, 326–334 (2008)
Article Google Scholar - Díaz, A. et al. Understanding the laminated layer of larval Echinococcus I: structure. Trends Parasitol. 27, 204–213 (2011)
Article Google Scholar - Hancock, K. et al. Characterization and cloning of GP50, a Taenia solium antigen diagnostic for cysticercosis. Mol. Biochem. Parasitol. 133, 115–124 (2004)
Article CAS Google Scholar - Heath, D. D., Jensen, O. & Lightowlers, M. W. Progress in control of hydatidosis using vaccination - a review of formulation and delivery of the vaccine and recommendations for practical use in control programmes. Acta Trop. 85, 133–143 (2003)
Article CAS Google Scholar - Zhang, G. et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54 (2012)
Article ADS CAS Google Scholar - Subjeck, J. R. & Repasky, E. A. Heat shock proteins and cancer therapy: the trail grows hotter!. Oncotarget 2, 433–434 (2011)
Article Google Scholar - Vargas-Parada, L., Solis, C. F. & Laclette, J. P. Heat shock and stress response of Taenia solium and T. crassiceps (Cestoda). Parasitology 122, 583–588 (2001)
Article CAS Google Scholar - Hemphill, A. et al. Echinococcus metacestodes as laboratory models for the screening of drugs against cestodes and trematodes. Parasitology 137, 569–587 (2010)
Article CAS Google Scholar - Brunetti, E. & White, A. C., Jr Cestode infestations: hydatid disease and cysticercosis. Infect. Dis. Clin. North Am. 26, 421–435 (2012)
Article Google Scholar - McVeigh, P. et al. Parasite neuropeptide biology: Seeding rational drug target selection? Int. J. Parasitol.. Drugs and Drug Res. 2, 76–91 (2012)
Article Google Scholar - Marks, N. J. & Maule, A. G. Neuropeptides in helminths: occurrence and distribution. Adv. Exp. Med. Biol. 692, 49–77 (2010)
Article CAS Google Scholar - Van Nassauw, L., Toovey, S., Van Op den Bosch, J., Timmermans, J. P. & Vercruysse, J. Schistosomicidal activity of the antimalarial drug, mefloquine, in _Schistosoma mansoni_-infected mice. Travel Med. Infect. Dis. 6, 253–258 (2008)
Article Google Scholar - Doudican, N., Rodriguez, A., Osman, I. & Orlow, S. J. Mebendazole induces apoptosis via Bcl-2 inactivation in chemoresistant melanoma cells. Mol. Cancer Res. 6, 1308–1315 (2008)
Article CAS Google Scholar - Grove, D. I. A History of Human Helminthology. 848 (CAB International, 1990)
Google Scholar - Spiliotis, M. & Brehm, K. Axenic in vitro cultivation of Echinococcus multilocularis metacestode vesicles and the generation of primary cell cultures. Methods Mol. Biol. 470, 245–262 (2009)
Article CAS Google Scholar - Spiliotis, M. et al. Echinococcus multilocularis primary cells: improved isolation, small-scale cultivation and RNA interference. Mol. Biochem. Parasitol. 174, 83–87 (2010)
Article CAS Google Scholar
Acknowledgements
We thank M. Dunn and OpGen for the E. multilocularis optical map; Roche for 20kb 454 libraries; R. Rance, M. Quail, D. Willey, N. Smerdon and K. Oliver for sequencing libraries at WTSI; T. D. Otto for bioinformatics expertise; R. Davies and Q. Lin for help with data release; A. Bateman, M. Punta, P. Cogghill and J. Minstry of Pfam; N. Rowlings from MEROPS database, J. Gough at SUPERFAMILY and C. Dessimoz for OMA; C. Seed and F. Jarero for laboratory assistance; J. Overington and B. Al-Lazikani; and B. Brejová for predicting genes with ExonHunter. P.D.O. and N.P.S. were supported in part by a BBSRC grant (BBG0038151) to P.D.O. P.D.O. and Ma.Z. were supported by a SynTax joint UK Research Council grant. The European Research Council supported P.W.H.H. and J.Paps. J.Parkinson and S.S.H. were supported by an operating grant from the Canadian Institute for Health Research (CIHR MOP#84556). G.S. was supported by FIRCA-NIH (grant TW008588) and the Universidad de la República, CSIC (grant CSIC 625). The E. multilocularis, E. granulosus and H. microstoma genome projects were funded by the Wellcome Trust through their core support of the Wellcome Trust Sanger Institute (grant 098051). K.B. was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG; BR2045/4-1). The Taenia solium Genome Project (IMPULSA 03) was supported by the Universidad Nacional Autonóma de México. The Taenia solium Genome Consortium thanks P. de la Torre, J. Yañez, P. Gaytán, S. Juárez and J. L. Fernández for technical support; L. Herrera-Estrella and LANGEBIO, CINVESTAV-Irapuato and C. B. Shoemaker for sequencing support and other advice; and J. Watanabe (deceased), S. Sugano and Y. Suzuki for the construction and sequencing of the full-length cDNA library.
Author information
Author notes
- Isheng J. Tsai, Magdalena Zarowiecki, Nancy Holroyd and Alejandro Garciarrubio: These authors contributed equally to this work.
Authors and Affiliations
- Parasite Genomics, Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SA, UK,
Isheng J. Tsai, Magdalena Zarowiecki, Nancy Holroyd, Alejandro Sanchez-Flores, Karen L. Brooks, Alan Tracey, Martin Aslett, Helen Beasley, Hayley M. Bennett, Richard Clark, Nishadi De Silva, Thomas Huckvale, Jacqueline A. Keane, Olivia Lambert, Sarah Nichol & Matthew Berriman - Division of Parasitology, Department of Infectious Disease, Faculty of Medicine, University of Miyazaki, Miyazaki 889-1692, Japan,
Isheng J. Tsai - Institute of Biotechnology, Universidad Nacional Autónoma de México, Cuernavaca, Morelos 62210, México,
Alejandro Garciarrubio, Alejandro Sanchez-Flores, Karel Estrada & Xavier Soberón - Institute of Biomedical Research, Universidad Nacional Autónoma de México, 04510 México D.F., México
Raúl J. Bobes, Gladis Fragoso, Edda Sciutto & Juan P. Laclette - State Key Laboratory of Veterinary Etiological Biology, Key Laboratory of Veterinary Parasitology of Gansu Province, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, No. 1 Xujiaping, Chengguan District, Lanzhou 730046, Gansu Province, China,
Jianping Cai, Junling Hou, Xuenong Luo, Yadong Zheng & Xuepeng Cai - Instituto de Microbiología y Parasitología Médica, Universidad de Buenos Aires-Consejo Nacional de Investigaciones Científicas y Tecnológicas (IMPaM, UBA-CONICET). Facultad de Medicina, Paraguay 2155, C1121ABG Buenos Aires, Argentina,
Federico Camicia, Marcela Cucher, Laura Kamenetzky, Natalia Macchiaroli & Mara Rosenzvit - Department of Biomedical Sciences, Iowa State University, Ames, Iowa 50011, USA,
Tim A. Day - Institute of Parasitology, Vetsuisse Faculty, University of Zürich, Winterthurerstrasse 266a, CH-8057 Zürich, Switzerland,
Peter Deplazes - Cátedra de Inmunologá, Facultad de Quámica, Universidad de la República. Avenida Alfredo Navarro 3051, piso 2, Montevideo, CP 11600, Uruguay,
Cecilia Fernández & Gustavo Salinas - Department of Zoology, University of Oxford, South Parks Road, Oxford OX1 3PS, UK,
Peter W. H. Holland & Jordi Paps - Beijing Institute of Genomics, Chinese Academy of Sciences, No.7 Beitucheng West Road, Chaoyang District, Beijing 100029, China,
Songnian Hu, Kan Liu & Yingfeng Luo - Department of Biochemistry & Molecular and Medical Genetics, University of Toronto, Program in Molecular Structure and Function, The Hospital for Sick Children, Toronto, Ontario M5G 1X8, Canada,
Stacy S. Hung & John Parkinson - University of Würzburg, Institute of Hygiene and Microbiology, D-97080 Würzburg, Germany
Ferenc Kiss, Uriel Koziol & Klaus Brehm - Department of Life Sciences, The Natural History Museum, Cromwell Road, London SW7 5BD, UK,
Natasha Pouchkina-Stantcheva, Nick Riddiford & Peter D. Olson - Department of Zoology, School of Natural Sciences and Regenerative Medicine Institute (REMEDI), National University of Ireland Galway, University Road, Galway, Ireland
Nick Riddiford - Department of Ecosystem and Public Health, Faculty of Veterinary Medicine, University of Calgary, Calgary T2N 4Z6, Canada
James D. Wasmuth - Institute of Parasitology, McGill University, 2111 Lakeshore Road, Ste Anne de Bellevue, Quebec H9X 3V9, Canada,
Mostafa Zamanian - Instituto Nacional de Medicina Genómica, Periférico Sur No. 4809 Col. Arenal Tepepan, Delegación Tlalpan, 14610 México, D.F., México,
Xavier Soberón - Institute of Biotechnology, Universidad Nacional Autónoma de México, Cuernavaca, Morelos 62210, México.,
Alejandro Garciarrubio, Alejandro Sánchez-Flores, Karel Estrada, Enrique Morett, Tobias Portillo & Xavier Soberón - Institute of Biomedical Research, Universidad Nacional Autónoma de México, 04510 México, D.F. Mexico.,
Raúl J. Bobes, Gladis Fragoso, Marco V. José, Edda Sciutto, Julio C. Carrero, Carlos Larralde, Jorge Morales-Montor, Jorge Limón-Lason & Juan P. Laclette - Genomic Sciences Center, Universidad Nacional Autónoma de México, Cuernavaca, Morelos 62210, Mexico.,
Miguel A. Cevallos & Víctor González - Instituto Nacional de Medicina Genómica, Periférico Sur No. 4809 Col. Arenal Tepepan, Delegación Tlalpan, 14610 México, D.F. México.,
Adrian Ochoa-Leyva & Xavier Soberón - School of Medicine, Universidad Nacional Autónoma de México, 04510 México, D.F. Mexico.,
Abraham Landa & Lucía Jiménez - School of Sciences, Universidad Nacional Autónoma de México, 04510 México, D.F. Mexico.,
Víctor Valdés
Authors
- Isheng J. Tsai
- Magdalena Zarowiecki
- Nancy Holroyd
- Alejandro Garciarrubio
- Alejandro Sanchez-Flores
- Karen L. Brooks
- Alan Tracey
- Raúl J. Bobes
- Gladis Fragoso
- Edda Sciutto
- Martin Aslett
- Helen Beasley
- Hayley M. Bennett
- Jianping Cai
- Federico Camicia
- Richard Clark
- Marcela Cucher
- Nishadi De Silva
- Tim A. Day
- Peter Deplazes
- Karel Estrada
- Cecilia Fernández
- Peter W. H. Holland
- Junling Hou
- Songnian Hu
- Thomas Huckvale
- Stacy S. Hung
- Laura Kamenetzky
- Jacqueline A. Keane
- Ferenc Kiss
- Uriel Koziol
- Olivia Lambert
- Kan Liu
- Xuenong Luo
- Yingfeng Luo
- Natalia Macchiaroli
- Sarah Nichol
- Jordi Paps
- John Parkinson
- Natasha Pouchkina-Stantcheva
- Nick Riddiford
- Mara Rosenzvit
- Gustavo Salinas
- James D. Wasmuth
- Mostafa Zamanian
- Yadong Zheng
- Xuepeng Cai
- Xavier Soberón
- Peter D. Olson
- Juan P. Laclette
- Klaus Brehm
- Matthew Berriman
Consortia
The Taenia solium Genome Consortium
- Alejandro Garciarrubio
- , Raúl J. Bobes
- , Gladis Fragoso
- , Alejandro Sánchez-Flores
- , Karel Estrada
- , Miguel A. Cevallos
- , Enrique Morett
- , Víctor González
- , Tobias Portillo
- , Adrian Ochoa-Leyva
- , Marco V. José
- , Edda Sciutto
- , Abraham Landa
- , Lucía Jiménez
- , Víctor Valdés
- , Julio C. Carrero
- , Carlos Larralde
- , Jorge Morales-Montor
- , Jorge Limón-Lason
- , Xavier Soberón
- & Juan P. Laclette
Contributions
I.J.T., Ma.Z., N.H. and M.B. Wrote the manuscript; M.B., K.B., J.P.L., X.S. and X.C. conceived and designed the project; N.H., M.B. and Ma.Z. coordinated the project. R.J.B., P.D., C.F., T.H., J.H., K.L., X.L., S.H., N.M., P.D.O., M.R., E.S., N.P.S., Y.Z. and K.B. prepared parasite material and nucleic acids; I.J.T., Ma.Z., A.G., K.E. and A.S.F. were involved in genome assembly; I.J.T., K.L.B., A.T., H.B., S.N., T.H., A.G. and K.E. were involved in genome assembly improvement; I.J.T., Ma.Z., A.S.F., X.S., K.L., J.H., A.G. and K.E. were involved in gene predictions; I.J.T., Ma.Z., K.L.B., A.T., H.B., O.L., S.N., R.C., R.J.B., G.F., E.S., X.S., J.P.L., K.L., J.H., A.G., K.E., S.H. and X.C. were involved in gene annotation; N.D.S., M.A., J.A.K., K.E., J.H., S.H., X.S. and Y.Z. were involved in data processing, computational and bioinformatics support; I.J.T. analysed genome structure, comparative genomics and ploidy; A.S.F., I.J.T. and Ma.Z. analysed gene structure; T.H. and H.M.B. experimentally validated micro-exons; A.G., K.B., F.K. and I.J.T. were involved with _trans_-splicing and polycistrons; I.J.T., S.S.H., J.Parkinson, G.S. and Ma.Z. examined metabolism and detoxification; P.W.H.H., J.Paps, N.R. and P.D.O. examined homeobox gene loss; K.B., I.J.T. and Ma.Z. examined stem cell specializations; Ma.Z. and J.D.W. examined domains; I.J.T., Ma.Z., C.F. and G.S. examined tapeworm-specific genes and expansions; Ma.Z. examined kinases and proteases; T.A.D. and Mo.Z. examined GPCRs; Mo.Z., T.A.D., U.K. and K.B. examined neuropeptides; M.R., L.K., F.C. and M.C. examined neuronal signalling; Ma.Z. examined drug targets; and G.S., M.R., C.F., K.B., P.W.H.H., P.D.O., A.G., R.J.B., G.F., E.S., X.S., J.P.L. and J.C. commented on the manuscript drafts.
Corresponding authors
Correspondence toJuan P. Laclette, Juan P. Laclette, Klaus Brehm or Matthew Berriman.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Text Sections 1-13, Supplementary Figures and Supplementary References – see contents for details. (PDF 7979 kb)
Supplementary Tables
This file contains Supplementary Tables 1-13. (XLSX 6151 kb)
Supplementary Tables
This file contains Supplementary Table 13.10. (XLSX 4407 kb)
PowerPoint slides
Rights and permissions
This article is distributed under the terms of the Creative Commons Attribution-Non-Commercial-Share Alike licence (http://creativecommons.org/licenses/by-nc-sa/3.0/), which permits distribution, and reproduction in any medium, provided the original author and source are credited. This licence does not permit commercial exploitation, and derivative works must be licensed under the same or similar licence.
About this article
Cite this article
Tsai, I., Zarowiecki, M., Holroyd, N. et al. The genomes of four tapeworm species reveal adaptations to parasitism.Nature 496, 57–63 (2013). https://doi.org/10.1038/nature12031
- Received: 09 November 2012
- Accepted: 21 February 2013
- Published: 13 March 2013
- Issue Date: 04 April 2013
- DOI: https://doi.org/10.1038/nature12031