Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase–deficient mice (original) (raw)
Main
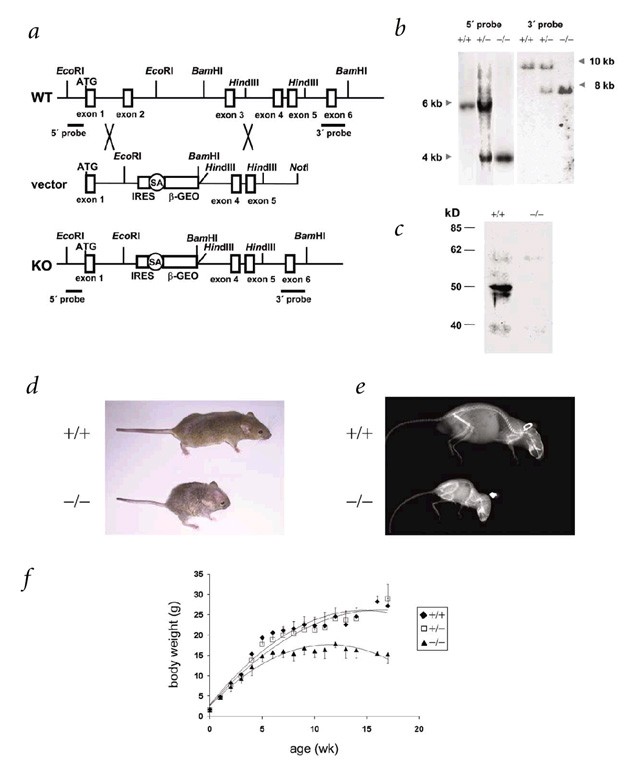
To clarify the function of Zmpste24, and to identify in vivo substrates targeted by this proteinase, we generated _Zmpste24_-null mice (Fig. 1_a_). After heterozygote intercrossing, we obtained _Zmpste24_-null, heterozygous and wildtype mice in the expected mendelian ratio. We verified homozygosity with respect to the mutated allele by Southern blot (Fig. 1_b_), and lack of Zmpste24 protein by western blot (Fig. 1_c_). Heterozygous mice were morphologically indistinguishable from their wildtype littermates. At birth, _Zmpste24_-null mice were also indistinguishable from their heterozygous or wildtype siblings and seemed healthy, until four weeks of age. Subsequently, null mice could be distinguished from wildtype littermates by size and weight. Within four to six weeks, their growth rate was reduced; by seven weeks, they stopped growing, despite normal feeding habits. At this stage, null mice were 30% smaller and had gained 40% less weight than their wildtype or heterozygous littermates (Fig. 1_d–f_). After two months of age, homozygous null mice began to progressively lose weight, showing an abnormal posture characterized by a hunched position and scoliosis (Fig. 1_e_). They became less mobile and began to slap and splay their hind paws while walking. When lifted by their tails, they reflexively overextended their hind limbs, trembled and were unable to bow upwards, unlike wildtype mice. In addition, most _Zmpste24_-null mice died prematurely, having an average lifespan of 20 weeks. We dissected animals between 14 and 20 weeks of age and found that the relative weight of the heart was greater in the mutants than in their littermate controls (0.53% ± 0.028% versus 0.44% ± 0.014%, mean ± s.e.m.; P<0.05). The relative weight of the kidneys was also higher in the _Zmpste24_-null mice than in controls (P<0.05).
Figure 1: Generation of _Zmpste24_-deficient mice.
a, Schematic representation of the wildtype Zmpste24 locus (WT), targeting vector and targeted allele (KO). Positions of restriction enzyme sites and probes used for Southern-blot analysis are shown. b, Southern-blot analysis of genomic DNA from three littermate progeny of Zmpste24 heterozygote crosses. Probing of _Eco_RI-digested DNA revealed fragments of 6 kb and 4 kb for wildtype and disrupted alleles, respectively. Probing of _Bam_HI-digested DNA revealed fragments of 10 kb and 8 kb for wildtype and disrupted alleles, respectively. c, Extracts of kidneys from wildtype and _Zmpste24_−/− mice were examined with a monoclonal antibody directed against the C terminus of Zmpste24. d, Photograph of two littermate progeny of a Zmpste24 heterozygote cross, at 3 mo. e, Radiograph of a _Zmpste24_−/− mouse at 3 mo, compared with a wildtype control. f, Cumulative plot of body weight versus age. Dots represent mean values, and error bars indicate s.e.m.
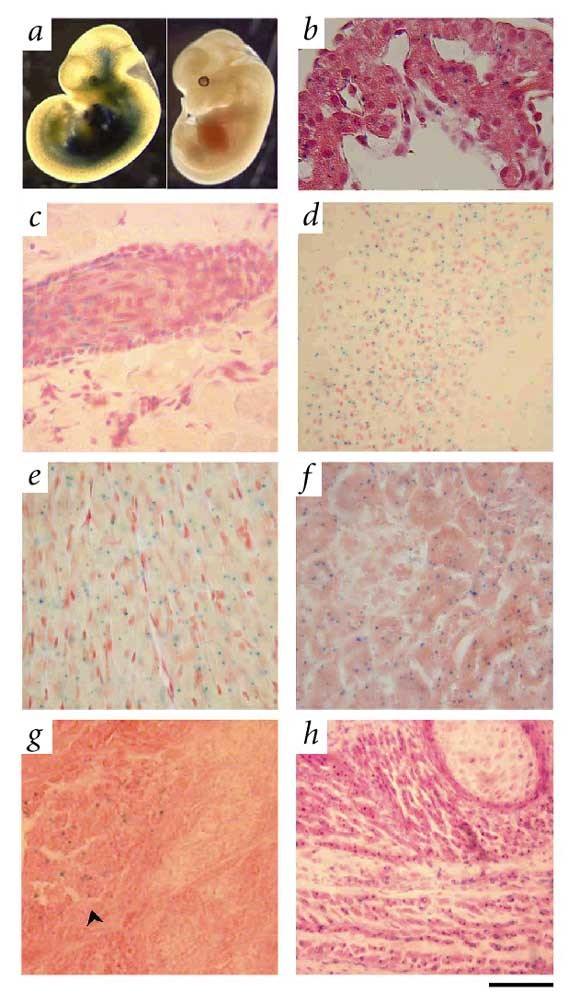
To analyze the molecular alterations underlying the observed abnormalities in _Zmpste24_-null mice, we first studied Zmpste24 expression during mouse development and in adult tissues. Northern-blot analysis identified a transcript of 3.5 kb in all tissues, indicating widespread expression of this gene (data not shown). To dissect more precisely the spatial and temporal expression pattern of Zmpste24 during mouse development, we took advantage of the _lacZ_-_neo_r fusion expressed under the control of the Zmpste24 promoter as a consequence of the targeting strategy. We saw extensive X-gal staining as early as embryonic day (E) 10.5, with the strongest activity being detected in the heart (Fig. 2_a_). We also carried out staining on tissue sections from mice at different stages of postnatal development, from 3 to 24 weeks of age (Fig. 2_b–h_). The intensity of the X-gal staining increased with age. We observed strong signals in cardiomyocytes, proximal and distal tubules and collecting ducts of kidney, neuronal cells of brain and the basal layer of epidermis, as well as hair follicles and fibroblasts of dermis. We also detected staining in pancreatic islets, smooth muscle cells of intestine, urinary bladder, blood vessels, seminiferous tubules of the testes, epithelial cells of the thyroid and skeletal muscle cells (Fig. 2 and data not shown).
Figure 2: Expression analysis of Zmpste24 during mouse development and in adult tissues.
a, Whole-mount staining of E10.5 embryos showing X-gal staining in heart, skin and central nervous system (left). b, Section of the same embryo showing X-gal staining in cardiomyocytes. c_–_h, X-gal staining of organ cryostat sections from an adult heterozygous mouse. c, Skin showing staining in keratinocytes of a hair follicle and in dermal fibroblasts. d, Hippocampus of the brain. Most of the staining localized to neuronal cells. e, Heart showing staining in cardiomyocytes. f, Kidney showing staining mainly in epithelial cells of proximal/distal tubules. g, Pancreas showing positive staining in endocrine (arrow), but not exocrine, tissue. h, Paravertebral muscle. Sections were counterstained with either eosin (b,f,g,h) or safranin (c_–_e). Scale bars represent 50 μm in b–g and 200 μm in h.
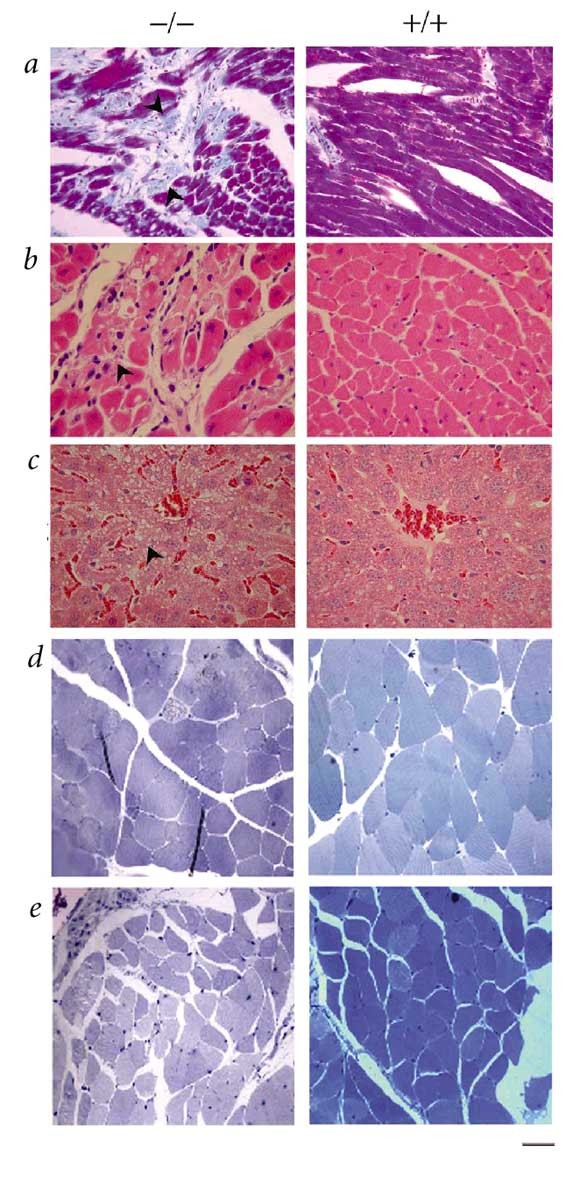
As Zmpste24 is highly expressed in the heart and kidney, we carried out serum and blood analyses to explore the possible causes of death. Blood glucose and serum triglyceride levels did not differ between _Zmpste24_-null mice and controls prior to the stage in which the mutants began to lose weight. Levels of both biochemical markers decreased in mutant animals that were losing weight, however (see Web Table A online). The serum levels of aspartate aminotransferase, creatine kinase, glutamate dehydrogenase and creatinine were normal in _Zmpste24_-null mice until 8 weeks of age. In end-stage mutants (approximately 20 weeks), however, we noticed a significant rise in serum aspartate aminotransferase, creatine kinase and glutamate dehydrogenase levels, whereas serum creatinine levels remained normal (see Web Table A online). These results suggest damage of the cardiac muscle. In agreement with serological data, we observed a significant thinning of the ventricular wall and, in some cases, a dilatation of both right and left ventricles. Microscopic analysis revealed foci of muscle degeneration, infiltration of inflammatory cells and interstitial fibrosis (Fig. 3_a_,b). We found an increased number of lipid droplets in hepatocytes surrounding the hepatic veins in liver from null mice, suggesting a chronic cardiac dysfunction (Fig. 3_c_). Taken together, these abnormalities observed in the heart suggest that dilated cardiomyopathy and heart failure probably caused the death of the _Zmpste24_-null mice.
Figure 3: Histology of heart, liver and muscle from _Zmpste24_-null (left) and wildtype (right) mice.
a, Gomori trichrome staining showing foci of muscle degeneration and fibrosis in mutant heart (left). Arrows indicate foci of fibrosis of the heart. Collagen was stained blue-green. b, Higher magnification (×400) of a heart section stained with hematoxylin and eosin showing muscle degeneration, lymphocytic infiltration, interstitial fibrosis and intracellular vesicles in the mutant heart (left). Arrow indicates a degenerating cardiac muscle and infiltration of lymphocytes. c, Histology of liver (×400) showing congestion and fatty degeneration of liver in the mutant mouse (left). Arrow indicates intracellular lipid accumulation. d,e, Semi-thin sections of quadriceps (d) and paravertebral (e) muscles at ×400 from mutant (left) and wildtype (right) mice. Scale bars represent 50 μm in a and c, and 25 μm in b,d and e.
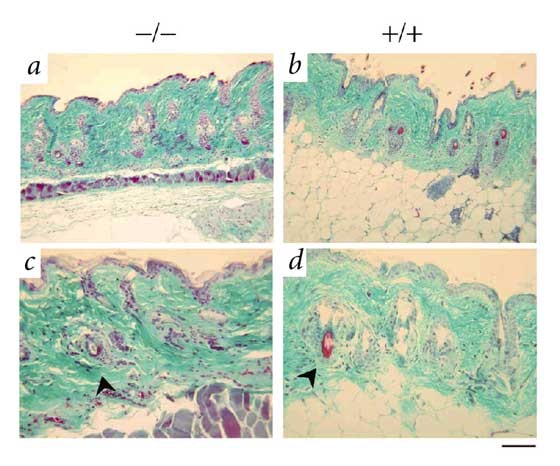
In addition to the heart pathology, muscular fibers of the para-vertebral region and the deltoid and quadriceps were dystrophic, showing a significant increase in abnormal small round fibers (Fig. 3_d–e_). Moreover, about one-third of the _Zmpste24_-null mice over 16 weeks of age started to lose their fur, whiskers and sometimes eyelashes. Microscopic analysis revealed that mutant mice at 16 weeks had completely lost the subcutaneous fat layer (Fig. 4). The epidermis and hair follicles were atrophic. There was an increased number of apoptotic bodies in the basal layer of the epidermis and in hair follicles, indicating the involvement of apoptosis in the pathogenesis of these skin-related lesions (Fig. 4). Notably, we saw thymic hypoplasia with loss of cortico-medullary demarcation in young null mice. Finally, there was a growth plate dysplasia in the distal femur and proximal tibia (data not shown). Mice deficient in Zmpste24 have previously been generated, but no phenotypic abnormalities or putative substrates targeted by this enzyme in mammalian cells were reported9. Apparently, these null mice were analyzed only during early development, which resulted in failure to detect the many phenotypes described here that are shown later in mouse development. Nevertheless, we cannot rule out the possibility that putative differences between those mice and the _Zmpste24_-null mice described here may be caused by the different targeting strategies used for inactivating the gene. The abnormalities that we observed in _Zmpste24_-null mice, including postnatal growth retardation, shortened lifespan, loss of fat layer and muscular dystrophy, are very similar to those developed by mice lacking A/C-type lamin4, a major protein component of the nuclear lamina5. Moreover, mutations in the gene encoding lamin A are responsible for a series of rare and seemingly unrelated human diseases that resemble the phenotype observed in the _Zmpste24_-null mice. These genetic disorders include dilated cardiomyopathy with conduction-system disease, Dunnigan-type familial partial lypodystrophy, Emery-Dreifuss muscular dystrophy and limb girdle muscular dystrophy (type 1B)6,7,8.
Figure 4: Masson trichrome staining of skin and subcutaneous tissue, showing lypodystrophy.
The _Zmpste24_-null mouse (left) shows complete loss of subcutaneous adipose tissue and severe atrophic epidermis and hair follicles (arrow), as compared with the wild type (right). a,b, Lower magnification (×200). c,d, Higher magnification (×400). Scale bars represent 100 μm in a and b, and 50 μm in c and d.
The finding that the _Zmpste24_-null phenotype in mice seemed to recapitulate the diverse pathological features associated with lamin A deficiencies in both human and mice led us to hypothesize that lamin A could be an in vivo substrate of Zmpste24. The lamin A protein is synthesized as a precursor that undergoes a complex maturation process, initiated by the farnesylation of a cysteine residue present in a conserved CaaX motif (C, cysteine; a, aliphatic; X, any residue) at the carboxy-terminal end of this protein. This step is followed by removal of the aaX tripeptide and methylation of the carboxyl group of the S-prenyl-cysteine10. These modifications are shared by other mammalian proteins, including the Ras family of oncoproteins and some heterotrimeric G proteins11. Farnesylated prelamin A undergoes a second endoproteolytic cleavage characteristic of this protein, however, resulting in the release of 15 amino acids from its C-terminal region, including the farnesylated cysteine residue12. The enzyme responsible for this specific endoproteolytic maturation step of the lamin A precursor has not been identified.
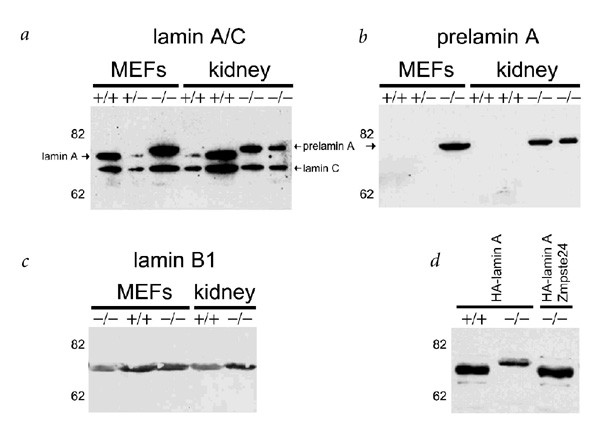
To evaluate whether Zmpste24 is involved in prelamin A processing, we first examined by western blot the maturation stage of this protein in cells and tissues from _Zmpste24_-null mice. Murine embryonic fibroblasts (MEFs) and tissue extracts from mutant mice lack the mature lamin A band (72 kD) present in wildtype extracts. Instead, we observed a band of the expected molecular mass for prelamin A (74 kD; Fig. 5_a_). We confirmed this finding by hybridizing the same blot with an antibody against the C-terminal tail, which is proteolytically removed during prelamin A maturation (Fig. 5_b_). High accumulation of prelamin A was seen in Zmpste24 −/− cell extracts, but was absent in wildtype controls. By contrast, we did not see molecular weight changes in either lamin C (Fig. 5_a_), a splice variant of lamin A without the CaaX box, or in lamin B1 (Fig. 5_c_), a CaaX-prenylated lamin that does not undergo the second proteolytic maturation event characteristic of lamin A. To demonstrate a direct role of Zmpste24 in the processing of prelamin A, we transfected wildtype and Zmpste24 −/− MEFs with an hemagglutinin-tagged murine prelamin A cDNA, alone or in combination with a Zmpste24 expression construct. When we analyzed the transfected cells by western blot, we detected bands of 72 kD and 74 kD in wildtype and _Zmpste24_-null mice, respectively, confirming the lack of prelamin A processing in the mutant cells (Fig. 5_d_). Co-transfection with the Zmpste24 expression plasmid resulted in the appearance of the 72 kD band in Zmpste24 −/− MEFs, indicating that re-expression of the proteinase is sufficient to revert the deficiency in prelamin A processing observed in these cells (Fig. 5_d_). These results indicate that Zmpste24 is necessary for the in vivo maturation of prelamin A.
Figure 5: Loss of Zmpste24 prevents prelamin A processing.
a–c, Extracts of MEFs and kidney from wildtype and mutant mice were probed with anti–lamin A/C (a), anti–prelamin A (b) or anti–lamin B1 antibodies (c). d, Wildtype and mutant MEFs were transfected with a hemagglutinin-tagged lamin A construct alone or together with a construct expressing Zmpste24 cDNA. Extracts of the transfected cells were analyzed with an anti-hemagglutinin monoclonal antibody.
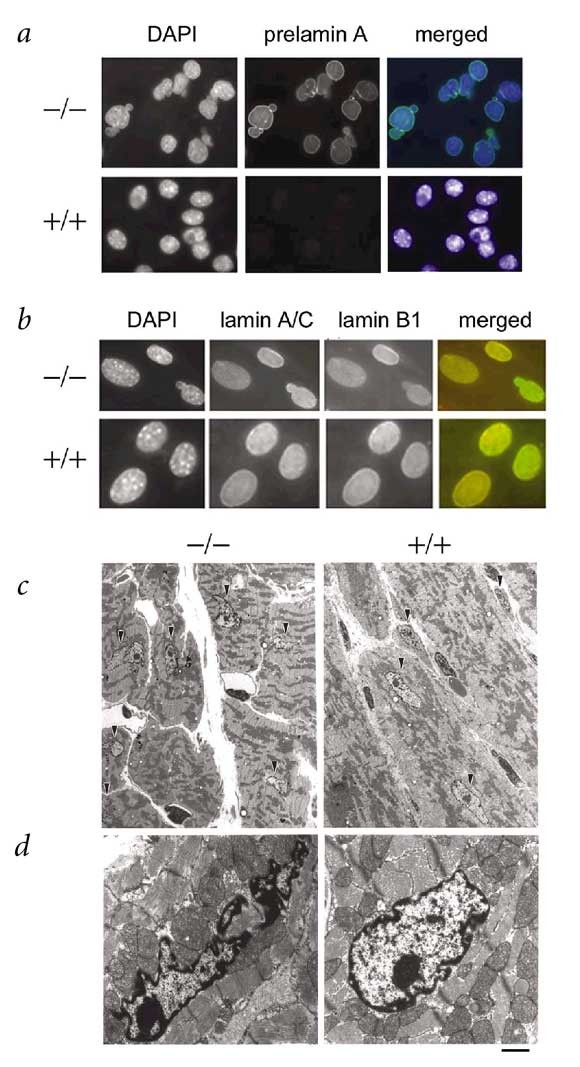
To examine whether the observed deficiency in prelamin A maturation causes nucleoskeleton abnormalities, we carried out immunofluorescence analysis of nuclear lamina components in MEFs and newborn fibroblasts of both wildtype and _Zmpste24_-null mice. Prelamin A was associated with the nuclear envelope in mutant cells (Fig. 6_a_). Co-localization of lamin B1 and lamin A/C confirmed this observation. Prelamin A, however, could not be readily detected in wildtype nuclei under the same conditions. By contrast, we did not observe differences in lamin B1 and emerin (two additional intermediate filament components of the nuclear lamina) between _Zmpste24_−/− cells and wildtype controls (Fig. 6_b_ and data not shown). We obtained similar results in wildtype and mutant MEFs transfected with a prelamin A cDNA that was tagged with hemagglutinin at its amino-terminal end (data not shown). These labeling experiments also revealed changes in the nuclear morphology of cultured mutant cells. Whereas nuclei of MEFs and neonatal fibroblasts are circular or ovoid, those of mutant cells are often of irregular shape and show herniation-like nuclear blebs (Fig. 6_a_,b). Electron microscopy analysis also confirmed the abnormalities in nuclear architecture of mutant cardiac muscle cells (83% abnormal shape in mutant nuclei versus 8% in wildtype; see Methods; Fig. 6_c_,d). Taken together, these results indicate that although uncleaved prelamin A is able to assemble normally in the nucleus during interphase, the integrity of the nuclear envelope becomes partially compromised in _Zmpste24_−/− cells. In addition, and consistent with the proposed role of Zmpste24 in prelamin A processing, immunofluorescence analysis of hemagglutinin-Zmpste24 transfected cells revealed that this protease localizes at the endoplasmic reticulum and at the nuclear envelope (data not shown).
Figure 6: Nuclear envelope architecture in wildtype and _Zmpste24_-deficient cells.
a,b, MEFs were analyzed by immunofluorescence with anti-prelamin A (a), or with anti-lamin A/C and anti-lamin B1 antibodies (b) and counterstained with DAPI. c, Electron microscope micrographs of cardiac muscle fibers and nuclei of cardiac muscle cells (arrowhead) showing irregular shape of the nuclei in mutant mice (left) compared with wildtype mice (right). d, Higher magnifications of the nuclei of cardiac muscle from mutant end-stage (left) and wildtype (right) mice. Scale bars represent 5 μm in c and 1 μm in d.
The links between prelamin A accumulation and tissue alterations could operate through two distinct but not mutually exclusive mechanisms. First, the altered structure of the nuclear lamina could affect directly mechanically stressed tissues, such as the muscle fibers of the heart and skeleton, where lamin B1 is not expressed13. In addition to this structural function, there is growing evidence that lamins are implicated in DNA replication, chromatin organization, regulation of transcription and cell division. These effects on nuclear dynamics may account for many of the clinical features and tissue-specific alterations observed in human laminopathies14,15,16. Our studies of _Zmpste24_-deficient mice confirm and extend these proposals. We provide further structural evidence of the occurrence of marked nuclear abnormalities associated with these diseases in mutant mice. The finding that proteins interacting with prelamin A and lamin A, like emerin, are incorporated into the nuclear lamina in mutant cells suggests that beyond structural alterations, downstream regulatory effects must also contribute to the observed phenotypes. Notably, the _Zmpste24_-null mice described here have a milder and later-acting phenotype than that observed in lamin A/C–deficient mice4. These _Zmpste24_-null mice can be considered a multiple knockout model, owing to the simultaneous deficiency of lamin A and lamin C and to the functional inactivation of emerin, which is mislocalized in lamin A/C–null cells4. Our Zmpste24 knockout model thus resembles more precisely the phenotype observed in individuals with hereditary laminopathies and may provide a useful tool to further dissect the mechanisms by which lamin alterations compromise muscular and adipocyte function, leading to these progressive and life-threatening diseases.
Methods
Gene targeting of Zmpste24.
We isolated PAC clones enclosing Zmpste24 from a 129 Sv library (HGMP Resource Centre, UK). This gene consists of 1,475 bp of coding sequence, organized in 10 exons distributed along a genomic segment of 38 kb (Fig. 1_a_). To create _Zmpste24_-null mice, we designed a targeting vector to replace exons 2 and 3 with a promoter-less IRES β-geo containing a splice acceptor sequence. Expression of the lacZ reporter gene was thus driven by the endogenous Zmpste24 promoter, and the Zmpste24 coding sequence was interrupted after codon 39. After electroporation of _Not_I-linearized targeting in 129 Ola embryonic stem cells, Southern-blot analysis of 120 neomycin-resistant colonies revealed homologous recombination in six independent clones. We used two of these heterozygote clones to generate chimeric mice. Five highly chimeric males were bred to C57BL/6 females; two gave germline transmission of the Zmpste24 targeted allele, as confirmed by PCR genotyping and Southern-blot analysis. Animal experimentation was done in accord with the guidelines of the Universidad de Oviedo and the Karolinska Institutet regarding the care and use of laboratory animals.
Histology, whole-mount staining and β-gal histochemistry.
We fixed samples with cold 4% paraformaldehyde in PBS, processed them into serial paraffin sections and stained them with hematoxylin and eosin or with the Masson's trichromic staining. We stained epoxy semithin muscle sections with toluidine blue following standard procedures. We carried out whole-mount embryo staining and histochemical analysis of β-galactosidase reporter activity in frozen sections as described17.
Electron microscopy.
We killed mice by cervical dislocation and immediately placed the heart in 2% glutaraldehyde and 0.5% paraformaldehyde in 0.1 M sodium cacodylate buffer containing 0.1 M sucrose and 3 mM CaCl2, pH 7.4. We cut small pieces from the myocardium and stored them in the fixative overnight at 4 °C. We rinsed samples in 0.15 M sodium cacodylate buffer containing 3 mM CaCl2 and post-fixed them in 2% OsO4 in 0.07 M sodium cacodylate buffer containing 3 mM CaCl2 for 2 h. Specimens were dehydrated and embedded in LX-112, and we examined them in a Tecnai 10 transmission electron microscope at 80 kV.
Evaluation of nuclear morphology in heart muscles.
We defined normal nuclei as spool-shaped or rounded, and pathological nuclei as irregular and more elongated. We randomly took electron micrographs and blindly selected counting areas at low magnification. We counted the number of 'pathological nuclei' at ×4400 and divided by the total number of nuclei in controls and knockouts, respectively. We analyzed a total of 60 wildtype and 104 mutant nuclei.
Expression constructs.
To produce hemagglutinin-lamin A (epitope-tagged at the N terminus), we subcloned a PCR fragment (_Bgl_II-X_ba_I) encoding mouse prelaminA in-frame into pcDNA3-HA (Invitrogen). Human FACE-1 cDNA was subcloned into pcDNA3.
Western blot and immunofluorescence analysis.
We separated in SDS-polyacrylamide gels total protein extracts homogenized in SDS lysis buffer containing Tris-HCl (60 mM, pH 6.8), 1% SDS, 8% glycerol. We transferred proteins to nitrocellulose and incubated them overnight at 4 °C with the different primary antibodies polyclonal anti–prelamin A (sc-6214, Santa Cruz Biotechnology), polyclonal anti–lamin B1 (sc-6217, Santa Cruz Biotechnology), monoclonal anti-lamin A/C (Manlac-1, provided by G. Morris), monoclonal anti–FACE-1 (Daiichi) and monoclonal anti-HA (Roche). We used a secondary antibody (rabbit anti-goat or goat anti-mouse) coupled to peroxidase for detection of the reaction with Supersignal Chemiluminiscent Substrate (Pierce). We derived MEFs from embryos at E13.5 as previously described18 and transiently transfected them with Fugene-6 (Roche). For immunofluorescence, we fixed cells in cold paraformaldehyde, rinsed them in PBS and permeabilized them with 0.2% Triton X-100. Finally, we incubated cells with the corresponding primary antibodies diluted in 15% FCS in PBS. The rhodamine and FITC-conjugated secondary antibodies were from Santa Cruz Biotechnology. We counterstained nuclei with DAPI.
References
- Freije, J.M.P. et al. Identification and chromosomal location of two human genes encoding enzymes potentially involved in proteolytic maturation of farnesylated proteins. Genomics 58, 270–280 (1999).
Article CAS Google Scholar - Tam, A. et al. Dual roles for Zmpste24p in yeast a-factor maturation: NH2-terminal proteolysis and COOH-terminal CAAX processing. J. Cell Biol. 142, 635–649 (1998).
Article CAS Google Scholar - Boyartchuck, V.L., Ashby, M.N. & Rine, J. Modulation of ras and a-factor function by carboxyl-terminal proteolysis. Science 275, 1796–1800 (1997).
Article Google Scholar - Sullivan, T. et al. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell. Biol. 147, 913–920 (1999).
Article CAS Google Scholar - Fisher, D.Z., Chaudhary, N. & Blobel, G. cDNA sequencing of nuclear lamins A and C reveals primary and secondary structural homology to intermediate filament proteins. Proc. Natl Acad. Sci. USA 83, 6450–6454 (1986).
Article CAS Google Scholar - Bonne, G. et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nature Genet. 21, 285–288 (1999).
Article CAS Google Scholar - Fatkin, D. et al. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N. Engl. J. Med. 341, 1715–1724 (1999).
Article CAS Google Scholar - Shackleton, S. et al. LMNA, encoding lamin A/C, is mutated in partial lipodystrophy. Nature Genet. 24, 153–156 (2000).
Article CAS Google Scholar - Leung, G.K. et al. Biochemical studies of Zmpste24-deficient mice. J. Biol. Chem. 276, 29051–29058 (2001).
Article CAS Google Scholar - Sinensky, M. et al. The processing pathway of prelamin A. J. Cell Sci. 107, 61–67 (1994).
CAS PubMed Google Scholar - Zhang, F.L. & Casey, P.J. Protein prenylation: molecular mechanisms and functional consequences. Annu. Rev. Biochem. 65, 241–269 (1996).
Article CAS Google Scholar - Weber, K., Plessmann, U. & Traub, P. Maturation of nuclear lamin A involves a specific carboxy-terminal trimming, which removes the polyisoprenylation site from the precursor; implications for the structure of the nuclear lamina. FEBS Lett. 257, 411–414 (1989).
Article CAS Google Scholar - Manilal, S. et al. Distribution of emerin and lamins in the heart and implications for Emery-Dreifuss muscular dystrophy. Hum. Mol. Genet. 8, 353–359 (1999).
Article CAS Google Scholar - Cohen, M., Lee, K.K., Wilson, K.L. & Gruenbaum, Y. Transcriptional repression, apoptosis, human disease and the functional evolution of the nuclear lamina. Trends Biochem. Sci. 26, 41–47 (2001).
Article CAS Google Scholar - Wilson, K.L., Zastrow, M.S. & Lee, K.L. Lamins and disease: insights into nuclear infrastructure. Cell 104, 647–650 (2001).
CAS PubMed Google Scholar - Morris, G.E. & Manilal, S. Heart to heart: from nuclear proteins to Emery-Dreifuss muscular dystrophy. Hum. Mol. Genet. 8, 1847–1850 (1999).
Article CAS Google Scholar - Hogan, B., Beddington, R., Costantini, F. & Lacy, E. Manipulating the Mouse Embryo: a Laboratory Manual (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 1994).
Google Scholar - Palmero, I. & Serrano, M. Induction of senescence by oncogenic Ras. Methods Enzymol. 333, 247–256 (2001).
Article CAS Google Scholar
Acknowledgements
We thank A.A. Ferrando, M. Serrano, D. Langenau, A. Fueyo, M. Balbín, G. Velasco, S. Cal, J.A. Uría and L.M. Sánchez for helpful comments; T. Look for support and advice; G. Morris, P. Crespo and K. Iwata for providing reagents; F. Alvarez for help in biochemical analysis and T. Sánchez, M. Fernández, U. Mikkonen and J. Wilbertz for excellent technical assistance. This work was supported by Comisión Interministerial de Ciencia y Tecnología, Fondos Plan FEDER, European Union, The Novo Nordisk Foundation, Swedish Medical Research Council and the Swedish Cancer Foundation. J.C. is the recipient of a fellowship from Gobierno Vasco (Spain). J.W. is a recipient of a Karolinska Institute Postdoc fellowship. The Instituto Universitario de Oncología is supported by Obra Social Cajastur-Asturias, Spain.
Author information
Author notes
- Alberto M. Pendás, Zhongjun Zhou and Juan Cadiñanos: These authors contributed equally to this work.
Authors and Affiliations
- Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, 33006, Spain
Alberto M. Pendás, Juan Cadiñanos, José M.P. Freije, Francisco Rodríguez & Carlos López-Otín - Division of Matrix Biology, Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Stockholm, Sweden
Zhongjun Zhou, Jianming Wang & Karl Tryggvason - Department of Pathology, Karolinska Institutet, Huddinge University Hospital, Huddinge, Sweden
Kjell Hultenby & Annika Wernerson - Servicio de Anatomía Patológica, Hospital Central de Asturias, Oviedo, Spain
Aurora Astudillo
Authors
- Alberto M. Pendás
You can also search for this author inPubMed Google Scholar - Zhongjun Zhou
You can also search for this author inPubMed Google Scholar - Juan Cadiñanos
You can also search for this author inPubMed Google Scholar - José M.P. Freije
You can also search for this author inPubMed Google Scholar - Jianming Wang
You can also search for this author inPubMed Google Scholar - Kjell Hultenby
You can also search for this author inPubMed Google Scholar - Aurora Astudillo
You can also search for this author inPubMed Google Scholar - Annika Wernerson
You can also search for this author inPubMed Google Scholar - Francisco Rodríguez
You can also search for this author inPubMed Google Scholar - Karl Tryggvason
You can also search for this author inPubMed Google Scholar - Carlos López-Otín
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toCarlos López-Otín.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Note: Supplementary information is available on the Nature Genetics website.
Supplementary information
Rights and permissions
About this article
Cite this article
Pendás, A., Zhou, Z., Cadiñanos, J. et al. Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase–deficient mice.Nat Genet 31, 94–99 (2002). https://doi.org/10.1038/ng871
- Received: 27 November 2001
- Accepted: 12 March 2002
- Published: 01 April 2002
- Issue Date: May 2002
- DOI: https://doi.org/10.1038/ng871