ChromHMM: automating chromatin-state discovery and characterization (original) (raw)
- Correspondence
- Published: 28 February 2012
Nature Methods volume 9, pages 215–216 (2012)Cite this article
- 47k Accesses
- 1520 Citations
- 20 Altmetric
- Metrics details
Subjects
To the Editor:
Chromatin-state annotation using combinations of chromatin modification patterns has emerged as a powerful approach for discovering regulatory regions and their cell type–specific activity patterns and for interpreting disease-association studies1,2,3,4,5. However, the computational challenge of learning chromatin-state models from large numbers of chromatin modification datasets in multiple cell types still requires extensive bioinformatics expertise. To address this challenge, we developed ChromHMM, an automated computational system for learning chromatin states, characterizing their biological functions and correlations with large-scale functional datasets and visualizing the resulting genome-wide maps of chromatin-state annotations.
This is a preview of subscription content, access via your institution
Relevant articles
Open Access articles citing this article.
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Additional access options:
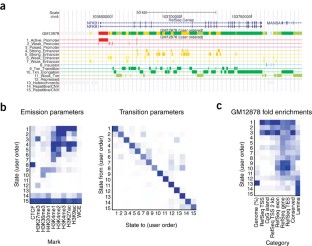
Figure 1: Sample outputs of ChromHMM.
References
- Day, N., Hemmaplardh, A., Thurman, R.E., Stamatoyannopoulos, J.A. & Noble, W.S. Bioinformatics 23, 1424–1426 (2007).
Article CAS Google Scholar - Ernst, J. & Kellis, M. Nat. Biotechnol. 28, 817–825 (2010).
Article CAS Google Scholar - Ernst, J. et al. Nature 473, 43–49 (2011).
Article CAS Google Scholar - Filion, G.J. et al. Cell 143, 212–224 (2010).
Article CAS Google Scholar - Roy, S. et al. Science 330, 1787–1797 (2010).
Article CAS Google Scholar - Kent, W.J. et al. Genome Res. 12, 996–1006 (2002).
Article CAS Google Scholar
Acknowledgements
We thank members of the Massachusetts Institute of Technology Computational Biology group and B.E. Bernstein for useful discussions related to this work. The work was supported by a US National Science Foundation postdoctoral fellowship 0905968 to J.E. and grants from the US National Institutes of Health (1-RC1-HG005334 to M.K. and 1 U54 HG004570 to B.E. Bernstein).
Author information
Author notes
- Jason Ernst
Present address: Present address: Department of Biological Chemistry, University of California Los Angeles, Los Angeles, California, USA.,
Authors and Affiliations
- Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA
Jason Ernst & Manolis Kellis - Broad Institute of Massachusetts Institute of Technology and Harvard, Cambridge, Massachusetts, USA
Jason Ernst & Manolis Kellis
Authors
- Jason Ernst
You can also search for this author inPubMed Google Scholar - Manolis Kellis
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toManolis Kellis.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Ernst, J., Kellis, M. ChromHMM: automating chromatin-state discovery and characterization.Nat Methods 9, 215–216 (2012). https://doi.org/10.1038/nmeth.1906
- Published: 28 February 2012
- Issue Date: March 2012
- DOI: https://doi.org/10.1038/nmeth.1906