Exploring long-range genome interactions using the WashU Epigenome Browser (original) (raw)
- Correspondence
- Published: 29 April 2013
- Rebecca F Lowdon1,
- Daofeng Li1,
- Heather A Lawson1,
- Pamela A F Madden2,
- Joseph F Costello3,4 &
- …
- Ting Wang1
Nature Methods volume 10, pages 375–376 (2013)Cite this article
- 6547 Accesses
- 156 Citations
- 10 Altmetric
- Metrics details
Subjects
To the Editor:
Eukaryotic chromosomes are a highly organized three-dimensional entity folded through a tightly regulated process1,2 with important functions that include bringing distal regulatory elements into the vicinity of their target gene promoters and arranging the chromosomes into distinct compartments3,4,5,6. Recent technological innovations, including chromosome conformation capture carbon copy (5C), Hi-C and chromatin interaction analysis by paired-end tag sequencing (ChIA-PET), have facilitated the discovery of chromosomal organization principles and folding architectures at unprecedented scales and resolution. Each technology also comes with corresponding computational tools7,8 to process and visualize its specific data type (Supplementary Note 1). However, visualizing and navigating long-range interaction data, as well as integrating these interactions with other epigenomics data, remains a much-desired capability and a daunting challenge9.
This is a preview of subscription content, access via your institution
Relevant articles
Open Access articles citing this article.
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Additional access options:
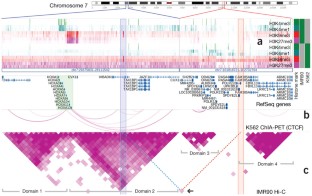
Figure 1: Two genomic regions as viewed in the WashU Epigenome Browser.
References
- Dekker, J. Science 319, 1793–1794 (2008).
Article CAS Google Scholar - Sanyal, A., Lajoie, B.R., Jain, G. & Dekker, J. Nature 489, 109–113 (2012).
Article CAS Google Scholar - Lieberman-Aiden, E. et al. Science 326, 289–293 (2009).
Article CAS Google Scholar - Handoko, L. et al. Nat. Genet. 43, 630–638 (2011).
Article CAS Google Scholar - Dixon, J.R. et al. Nature 485, 376–380 (2012).
Article CAS Google Scholar - Li, G. et al. Cell 148, 84–98 (2012).
Article CAS Google Scholar - Lajoie, B.R., van Berkum, N.L., Sanyal, A. & Dekker, J. Nat. Methods 6, 690–691 (2009).
Article CAS Google Scholar - Maunakea, A.K. et al. Nature 466, 253–257 (2010).
Article CAS Google Scholar - Stamatoyannopoulos, J.A. Genome Res. 22, 1602–1611 (2012).
Article CAS Google Scholar - Zhou, X. et al. Nat. Methods 8, 989–990 (2011).
Article CAS Google Scholar
Acknowledgements
We thank the laboratories of B. Ren, C.-L. Wei, Y. Ruan, J. Dekker; the UCSC Genome Browser team; and the ENCODE Consortium for providing long-range interaction data sets; and the Roadmap Epigenomics Consortium for providing epigenomics data sets. We thank many collaborators in the Roadmap Epigenomics Consortium who participated in software testing and provided helpful comments. We acknowledge support from the US National Institutes of Health (NIH) Roadmap Epigenomics Program, sponsored by the National Institute on Drug Abuse (NIDA) and the National Institute of Environmental Health Sciences (NIEHS). J.F.C. and T.W. are supported by NIH grant 5U01ES017154. X.Z. is supported by NIDA's R25 program DA027995. T.W. is supported in part by the March of Dimes Foundation, the Edward Jr. Mallinckrodt Foundation, NIH P50CA134254 and a grant from the Foundation for Barnes-Jewish Hospital.
Author information
Authors and Affiliations
- Department of Genetics, Center for Genome Sciences and Systems Biology, Washington University School of Medicine, St. Louis, Missouri, USA
Xin Zhou, Rebecca F Lowdon, Daofeng Li, Heather A Lawson & Ting Wang - Department of Psychiatry, Washington University in St. Louis, St. Louis, Missouri, USA
Pamela A F Madden - Department of Neurological Surgery, Brain Tumor Research Center, University of California, San Francisco, San Francisco, California, USA
Joseph F Costello - Helen Diller Family Comprehensive Cancer Center, University of California, San Francisco, San Francisco, California, USA
Joseph F Costello
Authors
- Xin Zhou
You can also search for this author inPubMed Google Scholar - Rebecca F Lowdon
You can also search for this author inPubMed Google Scholar - Daofeng Li
You can also search for this author inPubMed Google Scholar - Heather A Lawson
You can also search for this author inPubMed Google Scholar - Pamela A F Madden
You can also search for this author inPubMed Google Scholar - Joseph F Costello
You can also search for this author inPubMed Google Scholar - Ting Wang
You can also search for this author inPubMed Google Scholar
Corresponding authors
Correspondence toJoseph F Costello or Ting Wang.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Zhou, X., Lowdon, R., Li, D. et al. Exploring long-range genome interactions using the WashU Epigenome Browser.Nat Methods 10, 375–376 (2013). https://doi.org/10.1038/nmeth.2440
- Published: 29 April 2013
- Issue Date: May 2013
- DOI: https://doi.org/10.1038/nmeth.2440