NetGestalt: integrating multidimensional omics data over biological networks (original) (raw)
- Correspondence
- Published: 27 June 2013
Nature Methods volume 10, pages 597–598 (2013)Cite this article
- 7577 Accesses
- 58 Citations
- 18 Altmetric
- Metrics details
Subjects
To the Editor:
NetGestalt relies on a computational algorithm, available as the R package NetSAM (network seriation and modularization), for revealing hierarchical organization of biological networks (Supplementary Methods and Supplementary Fig. 1). Using protein-protein interaction networks from human and mouse as examples, we showed that NetSAM could identify biologically coherent modules at different scales and create functionally meaningful one-dimensional ordering of the genes (Supplementary Note 1, Supplementary Table 1 and Supplementary Fig. 2). Here we applied NetGestalt to a hierarchically and modularly organized human protein-protein interaction network based on interaction data from the Human Protein Reference Database (http://www.hprd.org/). We used two examples to demonstrate the features of NetGestalt and its potential in revealing important novel biological insights (Supplementary Note 1 and Supplementary Figs. 3–5).
This is a preview of subscription content, access via your institution
Relevant articles
Open Access articles citing this article.
Exome sequencing reveals a high genetic heterogeneity on familial Hirschsprung disease
- Berta Luzón-Toro
- , Hongsheng Gui
- … Salud Borrego
Scientific Reports Open Access 12 November 2015
Meta-analysis of human methylation data for evidence of sex-specific autosomal patterns
- Nina S McCarthy
- , Phillip E Melton
- … Alex W Hewitt
BMC Genomics Open Access 18 November 2014
Proteogenomic convergence for understanding cancer pathways and networks
- Emily S Boja
- & Henry Rodriguez
Clinical Proteomics Open Access 01 June 2014
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Additional access options:
Figure 1: One-dimensional layout of a network preserves the hierarchical modular organization information.
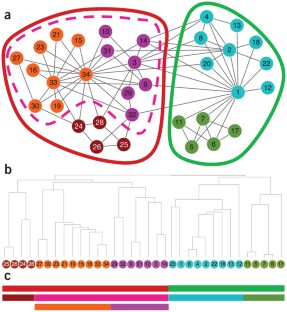
References
- Gehlenborg, N. et al. Nat. Methods 7, S56–S68 (2010).
Article CAS Google Scholar - Barabási, A.L. & Oltvai, Z.N. Nat. Rev. Genet. 5, 101–113 (2004).
Article Google Scholar - Cancer Genome Atlas Research Network. Nature 455, 1061–1068 (2008).
- Kent, W.J. et al. Genome Res. 12, 996–1006 (2002).
Article CAS Google Scholar
Acknowledgements
This work was supported by the US National Institutes of Health grants R01 GM088822, U24 CA159988 and P50 MH096972. This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University.
Author information
Author notes
- Zhiao Shi and Jing Wang: These authors contributed equally to this work.
Authors and Affiliations
- Advanced Computing Center for Research and Education, Vanderbilt University, Nashville, Tennessee, USA
Zhiao Shi - Department of Electrical Engineering and Computer Science, Vanderbilt University, Nashville, Tennessee, USA
Zhiao Shi - Department of Biomedical Informatics, Vanderbilt University, Nashville, Tennessee, USA
Jing Wang & Bing Zhang - Department of Cancer Biology, Vanderbilt University, Nashville, Tennessee, USA
Bing Zhang
Authors
- Zhiao Shi
- Jing Wang
- Bing Zhang
Corresponding author
Correspondence toBing Zhang.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5, Supplementary Tables 1–5, Supplementary Methods and Supplementary Notes 1 and 2 (PDF 6803 kb)
Rights and permissions
About this article
Cite this article
Shi, Z., Wang, J. & Zhang, B. NetGestalt: integrating multidimensional omics data over biological networks.Nat Methods 10, 597–598 (2013). https://doi.org/10.1038/nmeth.2517
- Published: 27 June 2013
- Issue Date: July 2013
- DOI: https://doi.org/10.1038/nmeth.2517