SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls (original) (raw)
Main
SARS-CoV-2 is the cause of COVID-192. This disease has been declared a pandemic by the World Health Organization (WHO), and is having severe effects on both individual lives and economies around the world. Infection with SARS-CoV-2 is characterized by a broad spectrum of clinical syndromes, which range from asymptomatic disease or mild influenza-like symptoms to severe pneumonia and acute respiratory distress syndrome3.
It is common to observe the ability of a single virus to cause widely differing pathological manifestations in humans. This is often due to multiple contributing factors including the size of the viral inoculum, the genetic background of patients and the presence of concomitant pathological conditions. Moreover, an established adaptive immunity towards closely related viruses4 or other microorganisms5 can reduce susceptibility6 or enhance disease severity7.
SARS-CoV-2 belongs to the Coronaviridae, a family of large RNA viruses that infect many animal species. Six other coronaviruses are known to infect humans. Four of them are endemically transmitted8 and cause the common cold (OC43, HKU1, 229E and NL63), while SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) have caused epidemics of severe pneumonia9. All of these coronaviruses trigger antibody and T cell responses in infected patients: however, antibody levels appear to wane faster than T cells. SARS-CoV-specific antibodies dropped below the limit of detection within 2 to 3 years10, whereas SARS-CoV-specific memory T cells have been detected even 11 years after SARS11. As the sequences of selected structural and non-structural proteins are highly conserved among different coronaviruses (for example, NSP7 and NSP13 are 100% and 99% identical, respectively, between SARS-CoV-2, SARS-CoV and the bat-associated bat-SL-CoVZXC2112), we investigated whether cross-reactive SARS-CoV-2-specific T cells are present in individuals who resolved SARS-CoV, and compared the responses with those present in individuals who recovered from SARS-CoV-2 infection. We also studied these T cells in individuals with no history of SARS or COVID-19 or of contact with patients with SARS-CoV-2. Collectively these individuals are hereafter referred to as individuals who were not exposed to SARS-CoV and SARS-CoV-2 (unexposed donors).
SARS-CoV-2-specific T cells in patients with COVID-19
SARS-CoV-2-specific T cells have just started to be characterized for patients with COVID-1913,14 and their potential protective role has been inferred from studies of patients who recovered from SARS15 and MERS16. To study SARS-CoV-2-specific T cells associated with viral clearance, we collected peripheral blood from 36 individuals after recovery from mild to severe COVID-19 (demographic, clinical and virological information is included in Extended Data Table 1) and studied the T cell response against selected structural (N) and non-structural proteins (NSP7 and NSP13 of ORF1) of the large SARS-CoV-2 proteome (Fig. 1a). We selected the N protein as it is one of the more-abundant structural proteins produced17 and has a high degree of homology between different betacoranaviruses18 (Extended Data Fig. 1).
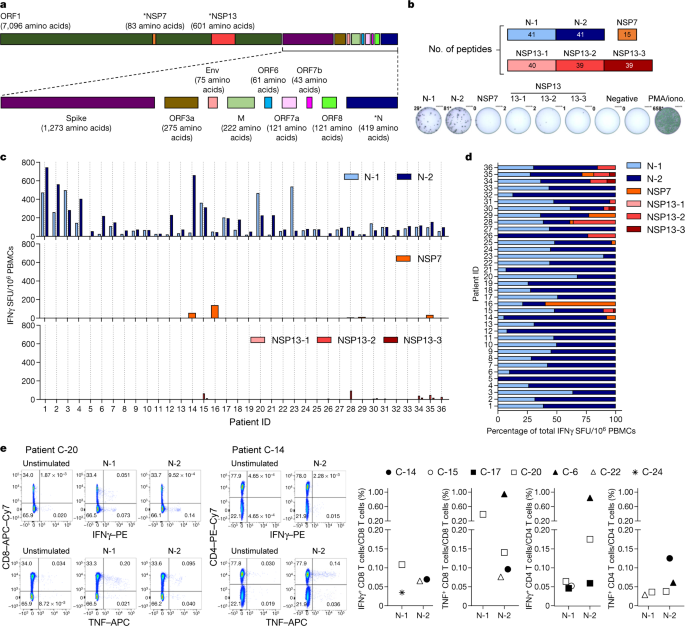
Fig. 1: SARS-CoV-2-specific responses in patients recovered from COVID-19.
a, SARS-CoV-2 proteome organization; analysed proteins are marked by an asterisk. b, The 15-mer peptides, which overlapped by 10 amino acids, comprising the N protein, NSP7 and NSP13 were split into 6 pools covering the N protein (N-1, N-2), NSP7 and NSP13 (NSP13-1, NSP13-2, NSP13-3). c, PBMCs of patients who recovered from COVID-19 (n = 36) were stimulated with the peptide pools or with phorbol 12-myristate 13-acetate (PMA) and ionomycin (iono) as a positive control. The frequency of spot-forming units (SFU) of IFNγ-secreting cells is shown. d, The composition of the SARS-CoV-2 response in each individual is shown as a percentage of the total detected response. N-1, light blue; N-2, dark blue; NSP7, orange; NSP13-1, light red; NSP13-2, red; NSP13-3, dark red. e, PBMCs were stimulated with the peptide pools covering the N protein (N-1, N-2) for 5 h and analysed by intracellular cytokine staining. Dot plots show examples of patients (2 out of 7) that had CD4 and/or CD8 T cells that produced IFNγ and/or TNF in response to stimulation with N-1 and/or N-2 peptides. The percentage of SARS-CoV-2 N-peptide-reactive CD4 and CD8 T cells in n = 7 individuals are shown (unstimulated controls were subtracted for each response).
NSP7 and NSP13 were selected for their complete homology between SARS-CoV, SARS-CoV-2 and other animal coronaviruses that belong to the betacoranavirus genus12 (Extended Data Fig. 2), and because they are representative of the ORF1a/b polyprotein that encodes the replicase–transcriptase complex19. This polyprotein is the first to be translated after infection with coronavirus and is essential for the subsequent transcription of the genomic and sub-genomic RNA species that encode the structural proteins19. We synthesized 216 15-mer peptides that overlapped by 10 amino acids and that covered the whole length of NSP7 (83 amino acids), NSP13 (601 amino acids) and N (422 amino acids) and split these peptides into five pools of approximately 40 peptides each (N-1, N-2, NSP13-1, NSP13-2 and NSP13-3) and a single pool of 15 peptides that spanned NSP7 (Fig. 1b). This unbiased method with overlapping peptides was used instead of bioinformatics selection of peptides, as the performance of such algorithms is often sub-optimal in Asian populations20.
Peripheral blood mononuclear cells (PBMCs) of 36 patients who recovered from COVID-19 were stimulated for 18 h with the different peptide pools and virus-specific responses were analysed by interferon-γ (IFNγ) ELISpot assay. In all individuals tested (36 out of 36), we detected IFNγ spots after stimulation with the pools of synthetic peptides that covered the N protein (Fig. 1c, d). In nearly all individuals, N-specific responses could be identified against multiple regions of the protein: 34 out of 36 individuals showed reactivity against the region that comprised amino acids 1–215 (N-1) and 36 out of 36 individuals showed reactivity against the region comprising amino acids 206–419 (N-2). By contrast, responses to NSP7 and NSP13 peptide pools were detected at very low levels in 12 out of 36 COVID-19-convalescent individuals tested.
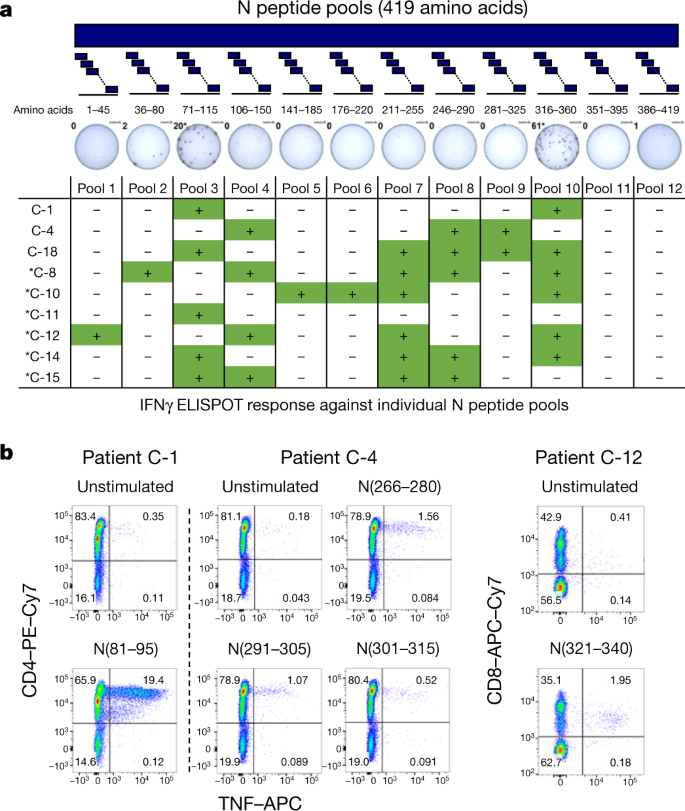
Direct ex vivo intracellular cytokine staining (ICS) was performed to confirm and define the N-specific IFNγ ELISpot response. Owing to their relative low frequency, N-specific T cells were more difficult to visualize by ICS than by ELISpot; however, a clear population of CD4 and/or CD8 T cells that produced IFNγ and/or TNF was detectable in seven out of nine analysed individuals (Fig. 1e and Extended Data Figs. 3, 4). Moreover, despite the small sample size, we could compare the frequency of SARS-CoV-2-specific IFNγ spots with the presence of virus-neutralizing antibodies, the duration of infection and disease severity and found no correlations (Extended Data Fig. 5). To confirm and further delineate the multi-specificity of the N-specific responses detected ex vivo in patients who recovered from COVID-19, we mapped the precise regions of the N protein that is able to activate IFNγ responses in nine individuals. We organized the 82 overlapping peptides that covered the entire N protein into small peptide pools (of 7–8 peptides) that were used to stimulate PBMCs either directly ex vivo or after an in vitro expansion protocol that has previously been used for patients with hepatitis B virus21 or SARS22. A schematic representation of the peptide pools is shown in Fig. 2a. We found that 8 out of 9 patients who recovered from COVID-19 had PBMCs that recognized multiple regions of the N protein of SARS-CoV-2 (Fig. 2a). Notably, we then defined single peptides that were able to activate T cells in seven patients. Using a peptide matrix strategy22, we first deconvolved the individual peptides that were responsible for the detected response by IFNγ ELISpot. Subsequently, we confirmed the identity of the single peptides by testing—using ICS—the ability of the peptides to activate CD4 or CD8 T cells (Table 1 and Fig. 2b). Table 1 summarizes the different T cell epitopes that were defined by both ELISpot and ICS for seven individuals who recovered from COVID-19. Notably, we observed that COVID-19-convalescent individuals developed T cells that were specific to regions that were also targeted by T cells from individuals who recovered from SARS. For example, the region of amino acids 101–120 of the N protein, which is a previously described CD4 T cell epitope in SARS-CoV-exposed individuals11,22, also stimulated CD4 T cells in two COVID-19-convalescent individuals. Similarly, the region of amino acids 321–340 of the N protein contained epitopes that triggered CD4 and CD8 T cells in patients who recovered from either COVID-19 or from SARS22. The finding that patients who recovered from COVID-19 and SARS can mount T cell responses against shared viral determinants suggests that previous SARS-CoV infection can induce T cells that are able to cross-react against SARS-CoV-2.
Fig. 2: SARS-CoV-2-specific T cells in COVID-19 convalescent individuals target multiple regions of the N protein.
a, PBMCs of 9 individuals who recovered from COVID-19 were stimulated with 12 different pools of 7–8 N peptides. The table shows IFNγ ELISpot responses against the individual N peptide pools. The asterisk denotes responses detected after in vitro expansion. b, After in vitro cell expansion, a peptide pool matrix strategy was used. T cells that reacted to distinct peptides were identified by IFNγ ELISpot and confirmed by ICS. Representative dot plots of 3 out of 7 patients are shown.
Table 1 SARS-CoV-2-specific T cell epitopes
SARS-CoV-2-specific T cells in patients with SARS
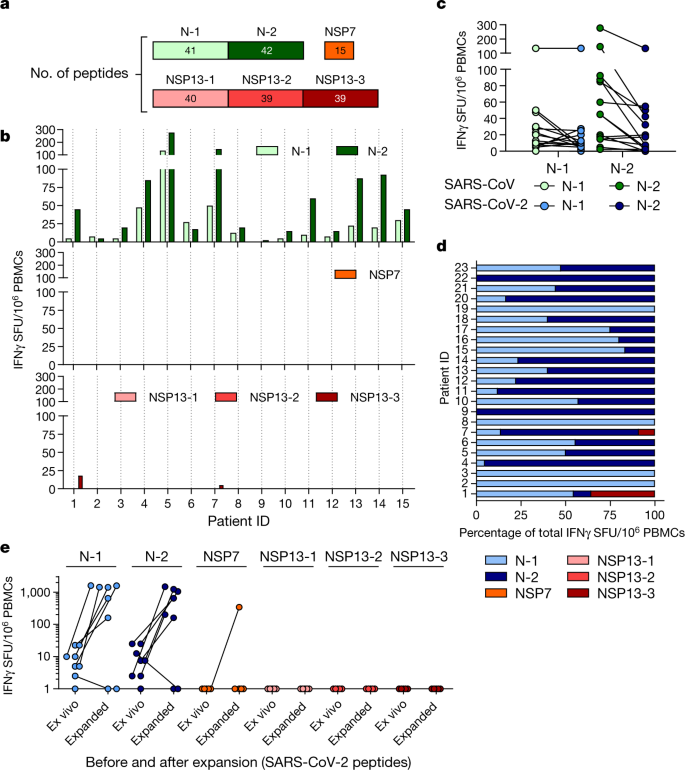
For the management of the current pandemic and for vaccine development against SARS-CoV-2, it is important to understand whether acquired immunity will be long-lasting. We have previously demonstrated that patients who recovered from SARS have T cells that are specific to epitopes within different SARS-CoV proteins that persist for 11 years after infection11. Here, we collected PBMCs 17 years after SARS-CoV infection and tested whether they still contained cells that were reactive against SARS-CoV and whether these had cross-reactive potential against SARS-CoV-2 peptides. PBMCs from individuals who had resolved a SARS-CoV infection (n = 15) were stimulated directly ex vivo with peptide pools that covered the N protein of SARS-CoV (N-1 and N-2), NSP7 and NSP13 (Fig. 3a). This revealed that 17 years after infection, IFNγ responses to SARS-CoV peptides were still present and were almost exclusively focused on the N protein rather than the NSP peptide pools (Fig. 3b). Subsequently, we tested whether the N peptides of SARS-CoV-2 (amino acid identity, 94%) induced IFNγ responses in PBMCs from individuals who resolved a SARS-CoV infection. Indeed, PBMCs from all 23 individuals tested reacted to N peptides from SARS-CoV-2 (Fig. 3c, d). To test whether these low-frequency responses in individuals who had recovered from SARS could expand after encountering the N protein of SARS-CoV-2, the quantity of IFNγ-producing cells that responded to the N, NSP7 and NSP13 proteins of SARS-CoV-2 was analysed after 10 days of cell culture in the presence of the relevant peptides. Seven out of eight individuals tested showed clear, robust expansion of N-reactive cells (Fig. 3e) and ICS confirmed that individuals who recovered from SARS had SARS-CoV N-reactive CD4 and CD8 memory T cells11 (Extended Data Fig. 6). In contrast to the response to the N peptides, we could not detect any cells that reacted to the peptide pools that covered NSP13 and only cells from one out of eight individuals reacted to NSP7 (Fig. 3e).
Fig. 3: SARS-CoV-2 cross-reactive responses are present in patients who recovered from SARS.
a, PBMCs isolated from 15 individuals who recovered from SARS 17 years ago were stimulated with SARS-CoV N, NSP7 and NSP13 peptide pools. b, Spot-forming units of IFNγ-secreting cells after overnight stimulation with the indicated peptide pools. c, PBMCs of 15 individuals who recovered from SARS were stimulated in parallel with peptide pools covering the N proteins of SARS-CoV and SARS-CoV-2, and the frequency of IFNγ-producing cells is shown. d, The composition of the SARS-CoV-2 response in each individual who recovered from SARS (n = 23) is shown as a percentage of the total detected response. N-1, light blue; N-2, dark blue; NSP7, orange; NSP13-1, light red; NSP13-2, red; NSP13-3, dark red. e, PBMCs of 8 individuals who recovered from SARS were stimulated with all peptides covering N, NSP7 and NSP13 of SARS-CoV-2 to detect cross-reactive responses. The numbers of cells that are reactive to the different peptide pools directly ex vivo and after in vitro expansion are shown.
Thus, SARS-CoV-2 N-specific T cells are part of the T cell repertoire of individuals with a history of SARS-CoV infection and these T cells are able to robustly expand after encountering N peptides of SARS-CoV-2. These findings demonstrate that virus-specific T cells induced by infection with betacoronaviruses are long-lasting, supporting the notion that patients with COVID-19 will develop long-term T cell immunity. Our findings also raise the possibility that long-lasting T cells generated after infection with related viruses may be able to protect against, or modify the pathology caused by, infection with SARS-CoV-2.
SARS-CoV-2-specific T cells in unexposed donors
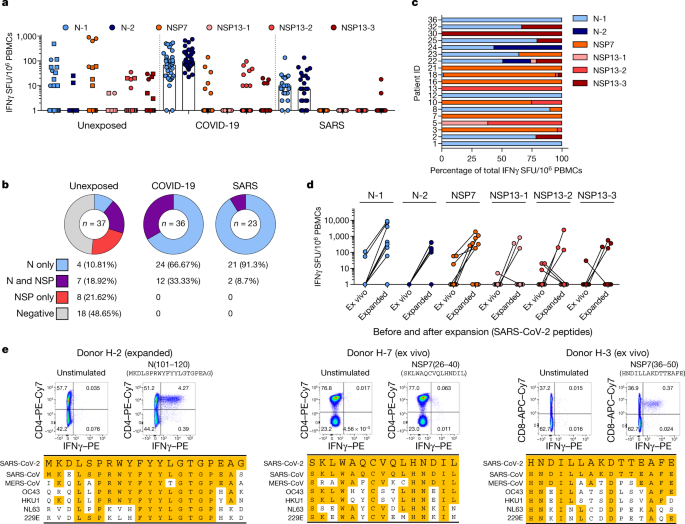
To explore this possibility, we tested N-, NSP7- and NSP13-peptide-reactive IFNγ responses in 37 donors who were not exposed to SARS-CoV and SARS-CoV-2. Donors were either sampled before July 2019 (n = 26) or were serologically negative for both SARS-CoV-2 neutralizing antibodies and SARS-CoV-2 N antibodies23 (n = 11). Different coronaviruses known to cause common colds in humans such as OC43, HKU1, NL63 and 229E present different degrees of amino acid homology with SARS-CoV-2 (Extended Data Fig. 1 and 2) and recent data have shown the presence of SARS-CoV-2 cross-reactive CD4 T cells (mainly specific to the spike protein) in donors who were not exposed to SARS-CoV-214. Notably, we detected SARS-CoV-2-specific IFNγ responses in 19 out of 37 unexposed donors (Fig. 4a, b). The cumulative proportion of all studied individuals who responded to peptides covering the N protein and the ORF1-encoded NSP7 and NSP13 proteins is shown in Fig. 4b. Unexposed donors showed a distinct pattern of reactivity; whereas individuals who recovered from COVID-19 and SARS reacted preferentially to N peptide pools (66% of individuals who recovered from COVID-19 and 91% of individuals who recovered from SARS responded to only the N peptide pools), the unexposed group showed a mixed response to the N protein or to NSP7 and NSP13 (Fig. 4a–c). In addition, whereas NSP peptides stimulated a dominant response in only 1 out of 59 individuals who had resolved COVID-19 or SARS, these peptides triggered dominant reactivity in 9 out of 19 unexposed donors with SARS-CoV-2-reactive cells (Fig. 4c and Extended Data Fig. 7). These SARS-CoV-2-reactive cells from unexposed donors had the capacity to expand after stimulation with SARS-CoV-2-specific peptides (Fig. 4d). We next delineated the SARS-CoV-2-specific response detected in unexposed donors in more detail. Characterization of the N-specific response in one donor (H-2) identified CD4 T cells that were reactive to an epitope within the region of amino acids 101–120 of the N protein. This epitope was also detected in patients who recovered from COVID-19 and SARS8,22 (Fig. 2b). This region has a high degree of homology to the sequences of the N protein of MERS-CoV, OC43 and HKU1 (Fig. 4e). In the same donor, we analysed PBMCs collected at multiple time points, demonstrating the persistence of the response to the 101–120 amino acid region of the N protein over 1 year (Extended Data Fig. 8a). In three other donors who were not exposed to SARS-CoV or SARS-CoV-2, we identified CD4 T cells specific to the region of amino acids 26–40 of NSP7 (SKLWAQCVQLHNDIL; donor H-7) and CD8 T cells specific to an epitope comprising the region of amino acids 36–50 of NSP7 (HNDILLAKDTTEAFE; H-3, H-21; Fig. 4e, Extended Data Fig. 8b).
Fig. 4: Immunodominance of SARS-CoV-2 responses in patients who recovered from COVID-19 and SARS, and in unexposed individuals.
a, PBMCs of individuals who were not exposed to SARS-CoV and SARS-CoV-2 (n = 37), recovered from SARS (n = 23) or COVID-19 (n = 36) were stimulated with peptide pools covering N (N-1, N-2), NSP7 and NSP13 (NSP13-1, NSP13-2, NSP13-3) of SARS-CoV-2 and analysed by ELISpot. The frequency of peptide-reactive cells is shown for each donor (dots or squares) and the bars represent the median frequency. Squares denote PBMC samples collected before July 2019. b, The percentage of individuals with N-specific, NSP7 and NSP13-specific responses, or N-, NSP7- and NSP13-specific responses in cohort. c, The composition of the SARS-CoV-2 response in each responding unexposed donor (n = 19) is shown as a percentage of the total detected response. N-1, light blue; N-2, dark blue; NSP7, orange; NSP13-1, light red; NSP13-2, red; NSP13-3, dark red. d, Frequency of SARS-CoV-2-reactive cells in 11 unexposed donors to the indicated peptide pools directly ex vivo and after a 10-day expansion. e, A peptide pool matrix strategy was used for three individuals who were not exposed to SARS-CoV and SARS-CoV-2. The identified T cell epitopes were confirmed by ICS, and the sequences were aligned to the corresponding sequence of all coronaviruses known to infect humans.
These latter two T cell specificities were of particular interest as the homology between the two protein regions of SARS-CoV, SARS-CoV-2 and other common cold coronaviruses (OC43, HKU1 NL63 and 229E) was minimal (Fig. 4e), especially for the CD8 T cell epitope. Indeed, the low-homology peptides that covered the sequences of the common cold coronaviruses failed to stimulate PBMCs from individuals with T cells responsive to amino acids 36–50 of NSP7 (Extended Data Fig. 8c). Even though we cannot exclude that some SARS-CoV-2-reactive T cells might be naive or induced by completely unrelated pathogens5, this finding suggests that unknown coronaviruses, possibly of animal origin, might induce cross-reactive SARS-CoV-2 T cells in the general population.
We further characterized the NSP7-specific CD4 and CD8 T cells that were present in the three unexposed individuals. The reactive T cells expanded efficiently in vitro and mainly produced either both IFNγ and TNF (CD8 T cells) or only IFNγ (CD4 T cells) (Extended Data Fig. 9a). We also determined that the CD8 T cells that were specific to amino acids 36–50 of NSP7 were HLA-B35-restricted and had an effector memory/terminal differentiated phenotype (CCR7−CD45RA+/−) (Extended Data Fig. 9b, c).
Conclusions
It is unclear why NSP7- and NSP13-specific T cells are detected and often dominant in unexposed donors, while representing a minor population in individuals who have recovered from SARS or COVID-19. It is, however, consistent with the findings of a previous study11, in which ORF1-specific T cells were preferentially detected in some donors who were not exposed to SARS-CoV-2 whereas T cells from individuals who had recovered from COVID-19 preferentially recognized structural proteins. Induction of virus-specific T cells in individuals who were exposed but uninfected has been demonstrated in other viral infections24,25,26. Theoretically, individuals exposed to coronaviruses might just prime ORF1-specific T cells, as the ORF1-encoded proteins are produced first in coronavirus-infected cells and are necessary for the formation of the viral replicase–transcriptase complex that is essential for the subsequent transcription of the viral genome, which then leads to the expression of various RNA species18. Therefore, ORF1-specific T cells could hypothetically abort viral production by lysing SARS-CoV-2-infected cells before the formation of mature virions. By contrast, in patients with COVID-19 and SARS, the N protein—which is abundantly produced in cells that secrete mature virions17—would be expected to preferentially boost N-specific T cells.
Notably, the ORF1 region contains domains that are highly conserved among many different coronaviruses9. The distribution of these viruses in different animal species might result in periodic human contact that induces ORF1-specific T cells with cross-reactive abilities against SARS-CoV-2. Understanding the distribution, frequency and protective capacity of pre-existing structural or non-structural protein-associated SARS-CoV-2 cross-reactive T cells could be important for the explanation of some of the differences in infection rates or pathology observed during this pandemic. T cells that are specific to viral proteins are protective in animal models of airway infections27,28, but the possible effects of pre-existing N- and/or ORF1-specific T cells onthe differential modulation of SARS-CoV-2 infection will have to be carefully evaluated.
Methods
Data reporting
No statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment.
Ethics statement
All donors provided written consent. The study was conducted in accordance with the Declaration of Helsinki and approved by the NUS Institutional Review Board (H-20-006) and the SingHealth Centralised Institutional Review Board (reference CIRB/F/2018/2387).
Human samples
Donors were recruited based on their clinical history of SARS-CoV or SARS-CoV-2 infection. Blood samples of patients who recovered from COVID-19 (n = 36) were obtained 2–28 days after PCR negativity and of patients who recovered from SARS (n = 23) 17 years after infection. Samples from healthy donors were either collected before June 2019 for studies of T cell function in viral diseases (n = 26), or in March–April 2020. All healthy donor samples tested negative for RBD-neutralizing antibodies and negative in an ELISA for N IgG (n = 11)19.
PBMC isolation
PBMCs were isolated by density-gradient centrifugation using Ficoll–Paque. Isolated PBMCs were either studied directly or cryopreserved and stored in liquid nitrogen until use in the assays.
Peptide pools
We synthesized 15-mer peptides that overlapped by 10 amino acids and spanned the entire protein sequence of the N, NSP7 and NSP13 proteins of SARS-CoV-2, as well as the N protein of SARS-CoV (GL Biochem Shanghai; see Supplementary Tables 1, 2). To stimulate PBMCs, the peptides were divided into 5 pools of about 40 peptides covering N (N-1, N-2) and NSP13 (NSP13-1, NSP13-2, NSP13-3) and one pool of 15 peptides covering NSP7. For single-peptide identification, peptides were organized in a matrix of 12 numeric and 7 alphabetical pools for N, and 4 numeric and 4 alphabetical pools for NSP7.
ELISpot assay
ELISpot plates (Millipore) were coated with human IFNγ antibody (1-D1K, Mabtech; 5 μg/ml) overnight at 4 °C. Then, 400,000 PBMCs were seeded per well and stimulated for 18 h with pools of SARS-CoV or SARS-CoV-2 peptides (2 μg/ml). For stimulation with peptide matrix pools or single peptides, a concentration of 5 μg/ml was used. Subsequently, the plates were developed with human biotinylated IFNγ detection antibody (7-B6-1, Mabtech; 1:2,000), followed by incubation with streptavidin-AP (Mabtech) and KPL BCIP/NBT Phosphatase Substrate (SeraCare). Spot forming units (SFU) were quantified with ImmunoSpot. To quantify positive peptide-specific responses, 2× mean spots of the unstimulated wells were subtracted from the peptide-stimulated wells, and the results expressed as SFU/106 PBMCs. We excluded the results if negative control wells had >30 SFU/106 PBMCs or positive control wells (phorbol 12-myristate 13-acetate/ionomycin) were negative.
Flow cytometry
PBMCs or expanded T cell lines were stimulated for 5 h at 37 °C with or without SARS-CoV or SARS-CoV-2 peptide pools (2 μg/ml) in the presence of 10 μg/ml brefeldin A (Sigma-Aldrich). Cells were stained with the yellow LIVE/DEAD fixable dead cell stain kit (Invitrogen) and anti-CD3 (clone SK7; 3:50), anti-CD4 (clone SK3; 3:50) and anti-CD8 (clone SK1; 3:50) antibodies. For analysis of the T cell differentiation status, cells were additionally stained with anti-CCR7 (clone 150503; 1:10) and anti-CD45RA (clone HI100; 1:10) antibodies. Cells were subsequently fixed and permeabilized using the Cytofix/Cytoperm kit (BD Biosciences-Pharmingen) and stained with anti-IFNγ (clone 25723, R&D Systems; 1:25) and anti-TNF (clone MAb11; 1:25) antibodies and analysed on a BD-LSR II FACS Scan. Data were analysed by FlowJo (Tree Star). Antibodies were purchased from BD Biosciences-Pharmingen unless otherwise stated.
Expanded T cell lines
T cell lines were generated as follows: 20% of PBMCs were pulsed with 10 μg/ml of the overlapping SARS-CoV-2 peptides (all pools combined) or single peptides for 1 h at 37 °C, washed and cocultured with the remaining cells in AIM-V medium (Gibco; Thermo Fisher Scientific) supplemented with 2% AB human serum (Gibco; Thermo Fisher Scientific). T cell lines were cultured for 10 days in the presence of 20 U/ml of recombinant IL-2 (R&D Systems).
HLA-restriction assay
The HLA type of healthy donor H-3 was determined and different Epstein–Barr virus (EBV)-transformed B cells lines with one common allele each were selected for presentation of peptide NSP7(36–50) (see below). B cells were pulsed with 10 μg/ml of the peptide for 1 h at 37 °C, washed three times and cocultured with the expanded T cell line at a ratio of 1:1 in the presence of 10 μg/ml brefeldin A (Sigma-Aldrich). Non-pulsed B cell lines served as a negative control for the detection of potential allogeneic responses and autologous peptide-pulsed cells served as a positive control. The HLA class I haplotype of the different B cell lines: CM780, A*24:02, A*33:03, B*58:01, B*55:02, Cw*07:02, Cw*03:02; WGP48, A*02:07, A*11:01, B*15:25, B*46:01, Cw*01:02, Cw*04:03; NP378, A*11:01, A*33:03, B*51:51, B*35:03, Cw*07:02, Cw*14:02; NgaBH, A*02:01, A*33:03, B*58:01, B*13:01, Cw*03:02.
Sequence alignment
Reference protein sequences for ORF1ab (accession numbers: QHD43415.1, NP_828849.2, YP_009047202.1, YP_009555238.1, YP_173236.1, YP_003766.2 and NP_073549.1) and the N protein (accession numbers: YP_009724397.2, AAP33707.1, YP_009047211.1, YP_009555245.1, YP_173242.1, YP_003771.1 and NP_073556.1) were downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/protein/). Sequences were aligned using the MUSCLE algorithm with default parameters and percentage identity was calculated in Geneious Prime 2020.1.2 (https://www.geneious.com). Alignment figures were made in Snapgene 5.1 (GSL Biotech).
Surrogate virus neutralization assay
A surrogate virus-neutralization test was used. Specifically, this test measures the quantity of anti-spike antibodies that block protein–protein interactions between the receptor-binding domain of the spike protein and the human ACE2 receptor using an ELISA-based assay[29](/articles/s41586-020-2550-z#ref-CR29 "Tan, C. W. et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2–spike protein–protein interaction. Nat. Biotechnol. https://doi.org/10.1038/s41587-020-0631-z
(2020).").Statistical analyses
All statistical analyses were performed in Prism (GraphPad Software); details are provided in the figure legends.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
Reference protein sequences for ORF1ab (accession numbers: QHD43415.1, NP_828849.2, YP_009047202.1, YP_009555238.1, YP_173236.1, YP_003766.2 and NP_073549.1) and the N protein (accession numbers: YP_009724397.2, AAP33707.1, YP_009047211.1, YP_009555245.1, YP_173242.1, YP_003771.1 and NP_073556.1) were downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/protein/). All data are available in the Article or the Supplementary Information. Source data are provided with this paper.
References
- Welsh, R. M. & Selin, L. K. No one is naive: the significance of heterologous T-cell immunity. Nat. Rev. Immunol. 2, 417–426 (2002).
Article CAS Google Scholar - Zhou, P. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020).
Article ADS CAS Google Scholar - Raoult, D., Zumla, A., Locatelli, F., Ippolito, G. & Kroemer, G. Coronavirus infections: epidemiological, clinical and immunological features and hypotheses. Cell Stress 4, 66–75 (2020).
Article CAS Google Scholar - Lim, M. Q. et al. Cross-Reactivity and anti-viral function of dengue capsid and NS3-specific memory T cells toward Zika virus. Front. Immunol. 9, 2225 (2018).
Article Google Scholar - Su, L. F., Kidd, B. A., Han, A., Kotzin, J. J. & Davis, M. M. Virus-specific CD4+ memory-phenotype T cells are abundant in unexposed adults. Immunity 38, 373–383 (2013).
Article CAS Google Scholar - Wen, J. et al. CD4+ T cells cross-reactive with dengue and Zika viruses protect against Zika virus infection. Cell Rep. 31, 107566 (2020).
Article CAS Google Scholar - Urbani, S. et al. Heterologous T cell immunity in severe hepatitis C virus infection. J. Exp. Med. 201, 675–680 (2005).
Article CAS Google Scholar - Nickbakhsh, S. et al. Epidemiology of seasonal coronaviruses: establishing the context for the emergence of coronavirus disease 2019. J. Infect. Dis. 222, 17–25 (2020).
Article Google Scholar - Cui, J., Li, F. & Shi, Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 17, 181–192 (2019).
Article CAS Google Scholar - Cao, W.-C., Liu, W., Zhang, P.-H., Zhang, F. & Richardus, J. H. Disappearance of antibodies to SARS-associated coronavirus after recovery. N. Engl. J. Med. 357, 1162–1163 (2007).
Article CAS Google Scholar - Ng, O.-W. et al. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine 34, 2008–2014 (2016).
Article CAS Google Scholar - Wu, A. et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 27, 325–328 (2020).
Article CAS Google Scholar - Ni, L. et al. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity 52, 971–977 (2020).
Article CAS Google Scholar - Grifoni, A. et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 181, 1489–1501 (2020).
Article CAS Google Scholar - Li, C. K.-F. et al. T cell responses to whole SARS coronavirus in humans. J. Immunol. 181, 5490–5500 (2008).
Article CAS Google Scholar - Zhao, J. et al. Recovery from the Middle East respiratory syndrome is associated with antibody and T-cell responses. Sci. Immunol. 2, eaan5393 (2017).
Article Google Scholar - Irigoyen, N. et al. High-resolution analysis of coronavirus gene expression by RNA sequencing and ribosome profiling. PLoS Pathog. 12, e1005473 (2016).
Article Google Scholar - de Wit, E., van Doremalen, N., Falzarano, D. & Munster, V. J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 14, 523–534 (2016).
Article Google Scholar - Knoops, K. et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6, e226 (2008).
Article Google Scholar - Rivino, L. et al. Defining CD8+ T cell determinants during human viral infection in populations of Asian ethnicity. J. Immunol. 191, 4010–4019 (2013).
Article CAS Google Scholar - Tan, A. T. et al. Host ethnicity and virus genotype shape the hepatitis B virus-specific T-cell repertoire. J. Virol. 82, 10986–10997 (2008).
Article CAS Google Scholar - Oh, H. L. J. et al. Engineering T cells specific for a dominant severe acute respiratory syndrome coronavirus CD8 T cell epitope. J. Virol. 85, 10464–10471 (2011).
Article CAS Google Scholar - Yong, S. E. F. et al. Connecting clusters of COVID-19: an epidemiological and serological investigation. Lancet Infect. Dis. 20, 809–815 (2020).
Article CAS Google Scholar - Rowland-Jones, S. L. et al. HIV-specific cytotoxic T-cell activity in an HIV-exposed but uninfected infant. Lancet 341, 860–861 (1993).
Article CAS Google Scholar - Park, S.-H. et al. Subinfectious hepatitis C virus exposures suppress T cell responses against subsequent acute infection. Nat. Med. 19, 1638–1642 (2013).
Article CAS Google Scholar - Werner, J. M., Abdalla, A., Gara, N., Ghany, M. G. & Rehermann, B. The hepatitis B vaccine protects re-exposed health care workers, but does not provide sterilizing immunity. Gastroenterology 145, 1026–1034 (2013).
Article CAS Google Scholar - Zhao, J. et al. Airway memory CD4+ T cells mediate protective immunity against emerging respiratory coronaviruses. Immunity 44, 1379–1391 (2016).
Article CAS Google Scholar - McKinstry, K. K. et al. Memory CD4+ T cells protect against influenza through multiple synergizing mechanisms. J. Clin. Invest. 122, 2847–2856 (2012).
Article CAS Google Scholar - Tan, C. W. et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2–spike protein–protein interaction. Nat. Biotechnol. https://doi.org/10.1038/s41587-020-0631-z (2020).
Acknowledgements
We thank M. K. Maini and S. Vasudevan for critical reading and editing of the manuscript. Grant support was provided by a Special NUHS COVID-19 Seed Grant Call, Project NUHSRO/2020/052/RO5+5/NUHS-COVID/6 (WBS R-571-000-077-733).
Author information
Author notes
- These authors contributed equally: Nina Le Bert, Anthony T. Tan
Authors and Affiliations
- Emerging Infectious Diseases Program, Duke-NUS Medical School, Singapore, Singapore
Nina Le Bert, Anthony T. Tan, Kamini Kunasegaran, Christine Y. L. Tham, Morteza Hafezi, Adeline Chia, Melissa Hui Yen Chng, Meiyin Lin, Nicole Tan, Martin Linster, Wan Ni Chia, Lin-Fa Wang, Eng Eong Ooi, Jenny Guek-Hong Low & Antonio Bertoletti - Institute of Molecular and Cell Biology (IMCB), A*STAR, Singapore, Singapore
Meiyin Lin & Yee-Joo Tan - National Centre of Infectious Diseases, Singapore, Singapore
Mark I-Cheng Chen - Department of Infectious Diseases, Singapore General Hospital, Singapore, Singapore
Shirin Kalimuddin & Jenny Guek-Hong Low - Department of Medicine, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Singapore
Paul Anantharajah Tambyah - Division of Infectious Disease, University Medicine Cluster, National University Hospital, Singapore, Singapore
Paul Anantharajah Tambyah - Department of Microbiology and Immunology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Singapore
Yee-Joo Tan - Singapore Immunology Network, A*STAR, Singapore, Singapore
Antonio Bertoletti
Authors
- Nina Le Bert
You can also search for this author inPubMed Google Scholar - Anthony T. Tan
You can also search for this author inPubMed Google Scholar - Kamini Kunasegaran
You can also search for this author inPubMed Google Scholar - Christine Y. L. Tham
You can also search for this author inPubMed Google Scholar - Morteza Hafezi
You can also search for this author inPubMed Google Scholar - Adeline Chia
You can also search for this author inPubMed Google Scholar - Melissa Hui Yen Chng
You can also search for this author inPubMed Google Scholar - Meiyin Lin
You can also search for this author inPubMed Google Scholar - Nicole Tan
You can also search for this author inPubMed Google Scholar - Martin Linster
You can also search for this author inPubMed Google Scholar - Wan Ni Chia
You can also search for this author inPubMed Google Scholar - Mark I-Cheng Chen
You can also search for this author inPubMed Google Scholar - Lin-Fa Wang
You can also search for this author inPubMed Google Scholar - Eng Eong Ooi
You can also search for this author inPubMed Google Scholar - Shirin Kalimuddin
You can also search for this author inPubMed Google Scholar - Paul Anantharajah Tambyah
You can also search for this author inPubMed Google Scholar - Jenny Guek-Hong Low
You can also search for this author inPubMed Google Scholar - Yee-Joo Tan
You can also search for this author inPubMed Google Scholar - Antonio Bertoletti
You can also search for this author inPubMed Google Scholar
Contributions
N.L.B. and A.T.T. designed all experiments and analysed all of the data, prepared the figures and edited the paper; K.K., C.Y.L.T., M.H., A.C., M.L. and N.T. performed ELISpots and intracellular cytokine staining, and generated short-term T cell lines; M.H.Y.C. and M.L. performed viral sequence homology and analysed data; W.N.C. and L.-F.W. carried out antibody testing; M.I-C.C., E.E.O., S.K., P.A.T., J.G.-H.L. and Y.-J.T. selected and recruited patients and analysed clinical data; Y.-J.T. provided funding and designed the study; AB designed and coordinated the study, provided funding, analysed the data and wrote the paper.
Corresponding author
Correspondence toAntonio Bertoletti.
Ethics declarations
Competing interests
A.B. is a cofounder of Lion TCR, a biotechnology company that develops T cell receptors for the treatment of virus-related diseases and cancers. All other authors have no competing interests related to the study.
Additional information
Peer review information Nature thanks Petter Brodin, Stanley Perlman and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Sequence alignment of the N protein from all types of human coronaviruses.
Amino acid sequences for the N protein were downloaded from the NCBI database and aligned using the MUSCLE algorithm. Conserved residues are highlighted in yellow and the degree of conservation is indicated by the coloured bars above.
Extended Data Fig. 2 Sequence alignment of the ORF1-encoded non-structural proteins NSP7 and NSP13 from all types of human coronaviruses.
Protein sequences for ORF1ab were downloaded from the NCBI database and aligned using the MUSCLE algorithm. The alignment for NSP7 and NSP13 is shown.
Extended Data Fig. 3 Flow cytometry gating strategy.
a, Forward scatter area (FSC-A) versus forward scatter height (FSC-H) density plot for doublet exclusion. b, Forward and side scatter (SSC-A) density plots to identify the lymphocyte population. c, Live T cells were gated based on CD3 expression and a live/dead discrimination dye. d, e, Only single expressing CD8 and CD4 T cells were Boolean gated (d) and used for IFNγ and/or TNF analysis (e).
Extended Data Fig. 4 IFNγ and TNF production profile of SARS-CoV-2-specific T cells of patients who recovered from COVID-19.
PBMCs from patients recovered from COVID-19 (n = 7) were stimulated with the peptide pools covering N (NP-1, NP-2) for 5 h and analysed by intracellular cytokine staining for IFNγ and TNF. Dot plots show examples of patients with CD8 (top) or CD4 (bottom) T cells that produced IFNγ and/or TNF in response to stimulation with N-1 or N-2 peptide pools. The bars show the respective single and double cytokine producing T cells as a proportion of the total detected response after stimulation with the corresponding N peptide pools in each patient who recovered from COVID-19.
Extended Data Fig. 5 Correlation analysis of SARS-CoV-2-specific IFNγ responses with the presence of virus-neutralizing antibodies, duration of infection and disease severity.
a, b, The magnitude of SARS-CoV-2-specific responses, as quantified by IFNγ ELISpot, against all (N, NSP7 and NSP13) SARS-CoV-2 proteins tested (left), N (middle) or NSP7 and NSP13 (right) was correlated with the level of virus-neutralizing antibodies assayed using a surrogate virus neutralization assay (a; n = 28) and the duration of SARS-CoV-2 PCR positivity (b; n = 34). The respective P values (two-tailed) and correlation coefficients (Spearman correlation) are indicated. Patients who present with mild (grey), moderate (orange) or severe (red) disease are indicated. c, Magnitude of SARS-CoV-2-specific responses stratified by mild (n = 26), moderate (n = 5) and severe (n = 5) disease. The bars represent the median magnitude of the response. Mild disease, with or without chest radiograph changes, not requiring oxygen supplement. Moderate disease, oxygen supplement less than 50%. Severe disease, oxygen supplement 50% or more or high-flow oxygen or intubation.
Extended Data Fig. 6 Analysis of SARS-CoV N response.
PBMCs of patient S-20 were expanded for 10 days and the frequency of T cells specific for the N-1 peptide pool were analysed by intracellular cytokine staining for IFNγ and TNF. Dot plots show CD8 and CD4 T cells that produced IFNγ and/or TNF in response to stimulation with the N-1 peptide pool.
Extended Data Fig. 7 Dominance of SARS-CoV-2 N, NSP7 and NSP13 responses in donors who recovered from COVID-19 or SARS as well as in unexposed individuals.
PBMCs from the respective individuals were stimulated with SARS-CoV-2 peptide pools as described in Fig. 1. The composition of the SARS-CoV-2 response is shown as a percentage of the total detected response in each group. N-1, light blue; N-2, dark blue; NSP7, orange; NSP13-1, light red; NSP13-2, red; NSP13-3, dark red. The proportion of individuals with NSP-dominant responses are illustrated in the pie charts.
Extended Data Fig. 8 Identification of SARS-CoV-2 epitopes in donors who were not exposed to SARS-CoV and SARS-CoV-2.
a, Longitudinal analysis of the SARS-CoV-2 N(101–120) response in individual H-2. PBMCs collected at the stated time points were stimulated with peptides spanning amino acids 101–120 of the N protein and assayed by IFNγ ELISpot. The frequencies of IFNγ SFU are shown. b, PBMCs were stimulated with the single peptides identified by the peptide matrix in parallel with the neighbouring peptides and assayed by IFNγ ELISpot. The amino acid residues are shown on the left; the frequency of IFNγ SFU on the right. Activating peptides are indicated in red and neighbouring peptides in black. c, PBMCs from individuals H-3 and H-21 were stimulated with the NSP7 peptide comprising amino acids 36–50 from SARS-CoV-2, MERS-CoV, OC43, HKU1, NL63 and 229E and analysed ex vivo by IFNγ ELISpot. A NSP7(36–50) T cell line expanded from individual H-3 was also tested with the corresponding peptides of other coronaviruses by IFNγ ELISpot. Amino acid sequences of the various peptides are shown in the table. Conserved amino acids are highlighted in yellow.
Extended Data Fig. 9 Characterization of SARS-CoV-2 NSP7-specific T cell responses in three individuals who were not exposed to SARS-CoV and SARS-CoV-2.
a, Dot plots show the frequency of IFNγ- and/or TNF-producing CD8 or CD4 T cells specific to the SARS-CoV-2 peptides directly ex vivo and after a 10-day expansion in three unexposed donors. b, The HLA class I haplotype of individual H-3 is shown in the table. HLA restriction of the NSP7(36–50)-specific T cells from this individual was deduced by co-culturing the T cells with NSP7(36–50)-peptide-pulsed EBV-transformed B cell lines that share the indicated HLA class I molecule (+). Activation of the NSP7(36–50)-specific T cells by autologous cells was achieved by the direct addition of the peptide and used as the positive control. c, The memory phenotype of CD8 T cells specific for NSP7(36–50) in individuals H-3 and H-21 were analysed ex vivo and shown in the dot plots. The frequencies of naive, effector memory, central memory and terminally differentiated NSP7(36–50)-specific CD8 T cells (red) are shown and density plots were overlaid on the total CD8 T cells (grey).
Extended Data Table 1 Donor characteristics
Supplementary information
Source data
Rights and permissions
About this article
Cite this article
Le Bert, N., Tan, A.T., Kunasegaran, K. et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls.Nature 584, 457–462 (2020). https://doi.org/10.1038/s41586-020-2550-z
- Received: 20 May 2020
- Accepted: 07 July 2020
- Published: 15 July 2020
- Issue Date: 20 August 2020
- DOI: https://doi.org/10.1038/s41586-020-2550-z