Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift (original) (raw)
Abstract
The recently emerged SARS-CoV-2 Omicron variant encodes 37 amino acid substitutions in the spike protein, 15 of which are in the receptor-binding domain (RBD), thereby raising concerns about the effectiveness of available vaccines and antibody-based therapeutics. Here we show that the Omicron RBD binds to human ACE2 with enhanced affinity, relative to the Wuhan-Hu-1 RBD, and binds to mouse ACE2. Marked reductions in neutralizing activity were observed against Omicron compared to the ancestral pseudovirus in plasma from convalescent individuals and from individuals who had been vaccinated against SARS-CoV-2, but this loss was less pronounced after a third dose of vaccine. Most monoclonal antibodies that are directed against the receptor-binding motif lost in vitro neutralizing activity against Omicron, with only 3 out of 29 monoclonal antibodies retaining unaltered potency, including the ACE2-mimicking S2K146 antibody[1](/articles/s41586-021-04386-2#ref-CR1 "Park, Y. J. et al. Antibody-mediated broad sarbecovirus neutralization through ACE2 molecular mimicry. Science https://doi.org/10.1126/science.abm8143
(2022)."). Furthermore, a fraction of broadly neutralizing sarbecovirus monoclonal antibodies neutralized Omicron through recognition of antigenic sites outside the receptor-binding motif, including sotrovimab[2](/articles/s41586-021-04386-2#ref-CR2 "Pinto, D. et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295 (2020)."), S2X259[3](/articles/s41586-021-04386-2#ref-CR3 "Tortorici, M. A. et al. Broad sarbecovirus neutralization by a human monoclonal antibody. Nature 597, 103–108 (2021).") and S2H97[4](/articles/s41586-021-04386-2#ref-CR4 "Starr, T. N. et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature 597, 97–102 (2021)."). The magnitude of Omicron-mediated immune evasion marks a major antigenic shift in SARS-CoV-2\. Broadly neutralizing monoclonal antibodies that recognize RBD epitopes that are conserved among SARS-CoV-2 variants and other sarbecoviruses may prove key to controlling the ongoing pandemic and future zoonotic spillovers.Similar content being viewed by others
Main
The evolution of RNA viruses can result in immune escape and modulation of binding to host receptors through the accumulation of mutations5. Previously emerged SARS-CoV-2 variants of concern (VOCs) have developed resistance to neutralizing antibodies, including some clinical antibodies that are used as therapeutics6,7,8. The B.1.351 (Beta) VOC showed the greatest magnitude of immune evasion from serum neutralizing antibodies6,7, whereas B.1.617.2 (Delta) quickly outcompeted all other circulating isolates through the acquisition of mutations that enhanced transmission and pathogenicity9,10,11 and eroded the neutralizing activity of antibody responses9.
The Omicron (B.1.1.529) variant was first detected in November 2021, was immediately declared to be a VOC by the World Health Organization (WHO) and quickly rose in frequency worldwide. The Omicron variant is substantially mutated compared to any previously described SARS-CoV-2 isolates, including 37 substitutions of residues in the spike protein in the predominant haplotype (Fig. 1a, Extended Data Figs. 1–4). Fifteen of the Omicron mutations are clustered in the RBD, which is the main target of neutralizing antibodies after infection or vaccination12,13, suggesting that Omicron might escape infection- and vaccine-elicited antibodies and therapeutic monoclonal antibodies. Nine of these mutations map to the receptor-binding motif (RBM), which is the RBD subdomain that directly interacts with the host receptor, ACE214.
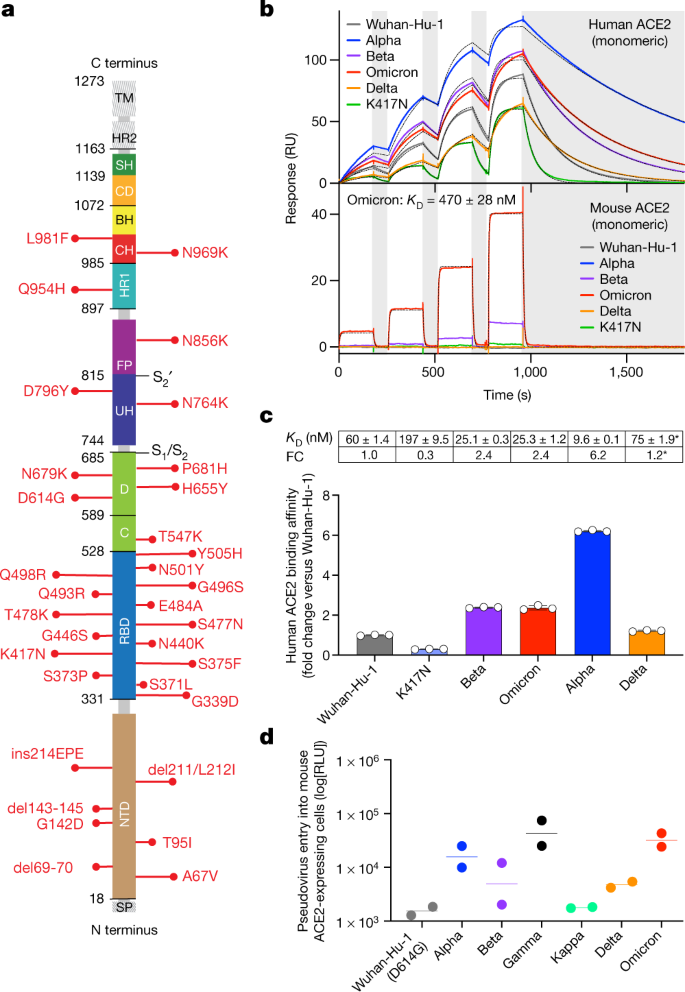
Fig. 1: The Omicron RBD shows increased binding to human ACE2 and gains binding to mouse ACE2.
a, Omicron mutations are shown in a primary structure of the SARS-CoV-2 spike protein, with domains and cleavage sites highlighted. BH, beta hairpin; C, C domain; CD, connector domain; CH, central helix; D, domain D; FP, fusion peptide;HR1/2, heptad repeat 1/2; SH, stem helix; SP, signal peptide; TM, transmembrane domain;UH, upstream helix. b, Single-cycle kinetics SPR analysis of ACE2 binding to six RBD variants. ACE2 is injected successively at 11, 33, 100 and 300 nM (human) or 33, 100, 300 and 900 nM (mouse). Dashed black curves show fits to a 1:1 binding model. White and grey stripes indicate association and dissociation phases, respectively. _K_D, dissociation constant; RU, response units. c, Quantification of human ACE2 binding data (mean ± s.d. of three replicates). Asterisks indicate that Delta was measured in a separate experiment with a different chip surface and capture tag. Delta fold change (FC) is calculated relative to the affinity of Wuhan-Hu-1 measured in parallel (91 ± 1.6 nM). d, Entry of Wuhan-Hu-1, Alpha, Beta, Delta, Gamma, Kappa and Omicron spike VSV pseudoviruses into mouse ACE2-expressing HEK293T cells. RLU, luciferase relative light units. Shown are two biological replicates (technical triplicates). Lines, geometric mean.
Preliminary reports indicate that the neutralizing activity of plasma from individuals who had received the Pfizer–BioNTech BNT162b2 vaccine is reduced against the Omicron variant15,[16](/articles/s41586-021-04386-2#ref-CR16 "Wilhelm, A. et al. Reduced neutralization of SARS-CoV-2 Omicron variant by vaccine sera and monoclonal antibodies. Preprint at https://doi.org/10.1101/2021.12.07.21267432
(2021)."), documenting a substantial—albeit not complete—escape from mRNA-vaccine-elicited neutralizing antibodies. Another report also shows that vaccine effectiveness against symptomatic disease induced by the Omicron variant is significantly lower than for the Delta variant[17](/articles/s41586-021-04386-2#ref-CR17 "Andrews, N. et al. Effectiveness of COVID-19 vaccines against the Omicron variant of concern. Preprint at
https://doi.org/10.1101/2021.12.14.21267615
(2021)."). The potential for booster doses to ameliorate this decline in neutralization is being investigated. In addition, the neutralizing activity of several therapeutic monoclonal antibodies appears to be decreased or abolished against Omicron[16](/articles/s41586-021-04386-2#ref-CR16 "Wilhelm, A. et al. Reduced neutralization of SARS-CoV-2 Omicron variant by vaccine sera and monoclonal antibodies. Preprint at
https://doi.org/10.1101/2021.12.07.21267432
(2021)."),[18](/articles/s41586-021-04386-2#ref-CR18 "Cao, Y. et al. B.1.1.529 escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature
https://doi.org/10.1038/s41586-021-04385-3
(2021).").To understand the consequences of the high number of mutations found in the Omicron spike protein, we used a pseudovirus assay to study receptor use and neutralization mediated by monoclonal and polyclonal antibodies, as well as surface plasmon resonance (SPR) to measure binding of the RBD to human and mouse ACE2 receptors.
Omicron RBD binds ACE2 with increased affinity
At present, 23 out of the 37 amino acid mutations in the Omicron spike protein have been individually observed previously in SARS-CoV-2 variants of interest, VOCs or other sarbecoviruses, whereas the remaining 14 substitutions have not to our knowledge been described before (Extended Data Fig. 5a). Analysis of the GISAID database indicates that there are rarely more than 10–15 Omicron spike mutations present in a given non-Omicron haplotype or Pango lineage (Extended Data Fig. 5b–d). Although we have not formally assessed the possibility of recombination events, persistent replication in immunocompromised individuals or inter-species ping-pong transmission5 are possible scenarios for the rapid accumulation of mutations that could have been selected on the basis of viral fitness and immune evasion.
Several of the Omicron RBD mutations are found at positions that are key contact sites with human ACE2, such as K417N, Q493R and G496S19. Except for N501Y, which increases ACE2-binding affinity by sixfold20,21, all other substitutions were shown by deep mutational scanning either to reduce binding or to have no effect on human ACE2 affinity when present individually22, resulting in an overall predicted decrease of binding affinity (Supplementary Table 1). However, we found that the Omicron RBD has a 2.4-fold increased binding affinity to human ACE2 (Fig. 1b, c, Extended Data Fig. 6a), suggesting epistasis of the full constellation of RBD mutations. It remains to be determined whether and how the spike mutations in Omicron may influence the dynamics of RBD opening, which may also affect the engagement of the RBD with ACE2.
The presence of the N501Y mutation has previously been reported to enable some SARS-CoV-2 VOCs to infect mice23. As Omicron contains the N501Y mutation, along with 14 other RBD mutations, we investigated whether the Omicron RBD binds mouse ACE2 using SPR (Fig. 1b, Extended Data Fig. 6). The Omicron RBD binds mouse ACE2 with a 1:1 binding affinity of 470 nM (Fig. 1b), whereas weak binding of the Beta RBD and very weak binding of the Alpha RBD to mouse ACE2 was observed (Fig. 1b, Extended Data Fig. 6b), consistent with previous reports23,24. Conversely, our assay did not detect any binding of the Wuhan-Hu-1, Delta or K417N RBDs to mouse ACE2. The enhanced binding of the Omicron RBD to mouse ACE2 is likely to be explained by theQ493R substitution, which is similar to the Q493K mutation isolated in mouse-adapted SARS-CoV-219. Our binding data correlate with ourobservation of Omicron spike protein-mediated but not Wuhan-Hu-1/G614 spike protein-mediated entry of VSV pseudoviruses into mouseACE2-expressing cells (Fig. 1d), as recently reported[25](/articles/s41586-021-04386-2#ref-CR25 "Hoffmann, M. et al. The Omicron variant is highly resistant against antibody-mediated neutralization—implications for control of the COVID-19 pandemic. Cell https://doi.org/10.1016/j.cell.2021.12.032
(2021)."). Collectively, these findings highlight the plasticity of the SARS-CoV-2 RBM, which in the case of the Omicron VOC acquired enhanced binding to human and mouse ACE2 orthologues, relative to other SARS-CoV-2 isolates. The influence of these findings on viral load and replication kinetics in humans and animal models remains to be evaluated, owing to the interplay of additional factors besides receptor binding. Preliminary data suggest that Omicron appears to be attenuated in some laboratory mouse strains (M.S.D., personal communication) and that it replicates less efficiently in human lung tissue as compared to Delta[26](/articles/s41586-021-04386-2#ref-CR26 "Nicholls, J. & Chan Chi-wai, M. HKUMed finds Omicron SARS-CoV-2 can infect faster and better than Delta in human bronchus but with less severe infection in lung.
www.med.hku.hk/en/news/press/20211215-omicron-sars-cov-2-infection
(2021).").Extent of Omicron escape from plasma antibodies
To investigate the magnitude of immune evasion that is mediated by the 37 mutations present in the Omicron spike protein, we used Wuhan-Hu-1 and Omicron spike VSV pseudoviruses and compared plasma neutralizing activity in different cohorts of convalescent individuals (that is, individuals who had recovered from COVID-19) or individuals who had been vaccinated with six major COVID-19 vaccines (mRNA-1273, BNT162b2, AZD1222, Ad26.COV2.S, Sputnik V and BBIBP-CorV) (Fig. 2, Supplementary Figs. 1–3, Extended Data Table 1).
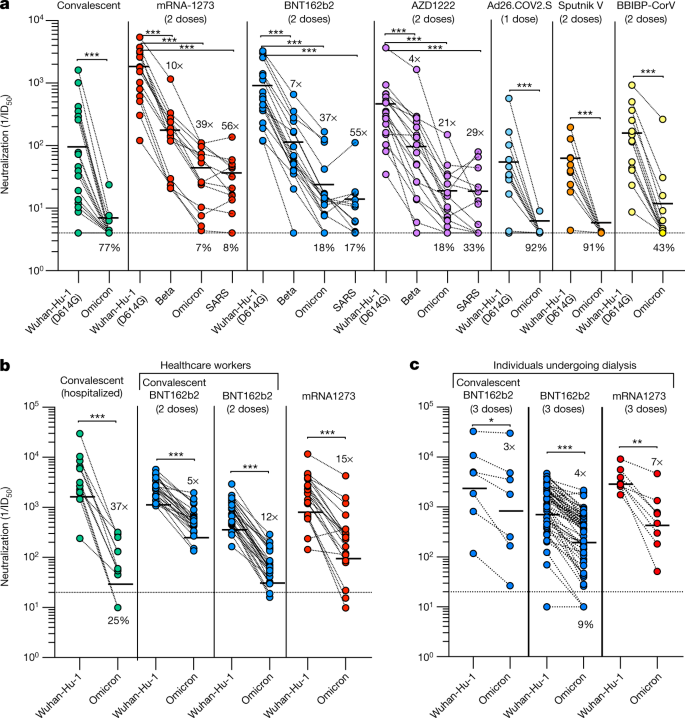
Fig. 2: Neutralization of Omicron SARS-CoV-2 VSV pseudovirus by plasma from convalescent and vaccinated individuals.
Plasma neutralizing activity in convalescent or vaccinated individuals (mRNA-1273, BNT162b2, AZD1222, Ad26.COV2.S (single dose), Sputnik V and BBIBP-CorV). a, Pairwise neutralizing antibody titres (half-maximum inhibitory dose; ID50) against Wuhan-Hu-1 (D614G), Beta and Omicron VOCs, and SARS-CoV. Vero E6-TMPRSS2 cells were used as target cells. Data are the geometric mean of n = 3 biologically independent experiments except for SARS-CoV for which n = 1. b, Pairwise neutralizing antibody titres of plasma (ID50) against Wuhan-Hu-1 and Omicron. Data are the geometric mean of n = 2 biologically independent experiments. c, Plasma neutralizing activity in individuals undergoing dialysis who received three doses of either the BNT162b2 or mRNA-1273. Pairwise neutralizing antibody titres of plasma (ID50) against Wuhan-Hu-1 and Omicron. One representative experiment out of two is shown. Vero E6 cells were used as target cells in b, c, Line, geometric mean of 1/ID50 titres. Shown is the percentage of samples that lost detectable neutralization against Omicron or SARS-CoV, excluding samples with 1/ID50 below the limit of detection. The demographics of enrolled donors are provided in Extended Data Table 1. Statistical significance is set as P < 0.05 and P values are indicated with asterisks (*P = 0.033; **P = 0.002; ***P < 0.001), using a paired two-sided _t_-test (Wilcoxon rank test).
Convalescent individuals and individuals who had been vaccinated with Ad26.COV2.S (single dose), Sputnik V or BBIBP-CorV had no detectable neutralizing activity against Omicron, with only one convalescent subject and three BBIBP-CorV vaccinees exhibiting ID50 values above 10 (Fig. 2a). Individuals who were immunized with mRNA-1273, BNT162b2 and AZD1222 had more potent neutralizing activity against Wuhan-Hu-1 and retained detectable neutralization against Omicron, with decreases of 39-, 37- and 21-fold, respectively (Fig. 2a). The dampening of neutralizing activity against Omicron was comparable to that observed against SARS-CoV, a virus that differs from Wuhan-Hu-1 by 52 residues in the RBD. Reductions of neutralization potency were less pronounced in vaccinated individuals who had been previously infected (fivefold; Fig. 2b) and in individuals undergoing dialysis (fourfold; Fig. 2c) who were boosted with a third mRNA vaccine dose. In the same cohort of individuals undergoing dialysis, the levels of antibodies that neutralize the vaccine-matched Wuhan-Hu-1 strain were found to be low (less than 1/100) or undetectable in 44% of individuals after the second dose of mRNA vaccine[27](/articles/s41586-021-04386-2#ref-CR27 "Bassi, J. et al. Poor neutralization and rapid decay of antibodies to SARS-CoV-2 variants in vaccinated dialysis patients. Preprint at https://doi.org/10.1101/2021.10.05.21264054
(2021).").Collectively, these findings provide evidence of a substantial reduction in plasma neutralizing activity against Omicron as compared to the ancestral virus, with neutralizing activity probably falling below the protective threshold in several cases28. Our data further indicate that multiple exposures to the ancestral virus through infection or vaccination result in the production of antibodies that can neutralize divergent viruses, such as Omicron or even SARS-CoV, as a consequence of affinity maturation or epitope masking by immune-dominant RBM antibodies28,[29](#ref-CR29 "Stamatatos, L. et al. mRNA vaccination boosts cross-variant neutralizing antibodies elicited by SARS-CoV-2 infection. Science https://doi.org/10.1126/science.abg9175
(2021)."),[30](/articles/s41586-021-04386-2#ref-CR30 "Bergstrom, J. J., Xu, H. & Heyman, B. Epitope-specific suppression of IgG responses by passively administered specific IgG: evidence of epitope masking. Front. Immunol. 8, 238 (2017).").Broadly neutralizing antibodies inhibit Omicron
Neutralizing monoclonal antibodies with demonstrated in vivo efficacy in the prevention or treatment of COVID-1931,32,33,34,35,36,37 can be divided into two groups on the basis of whether they do or do not block the binding of the spike protein to ACE2. Of the eight currently authorized or approved monoclonal antibodies, seven (LY-CoV555, LY-CoV016, REGN10987, REGN10933, COV2-2130, COV2-2196 and CT-P59; all synthesized on the basis of publicly available sequences) block the binding of spike protein to ACE2 and are often used as two-antibody cocktails8. They bind to epitopes that overlap with the RBM (Fig. 3a), which is structurally and evolutionary plastic38, as shown by the accumulation of mutations throughout the pandemic and the genetic diversity of this subdomain among ACE2-using sarbecoviruses[39](/articles/s41586-021-04386-2#ref-CR39 "Starr, T. N. et al. ACE2 binding is an ancestral and evolvable trait of sarbecoviruses. Preprint at https://doi.org/10.1101/2021.07.17.452804
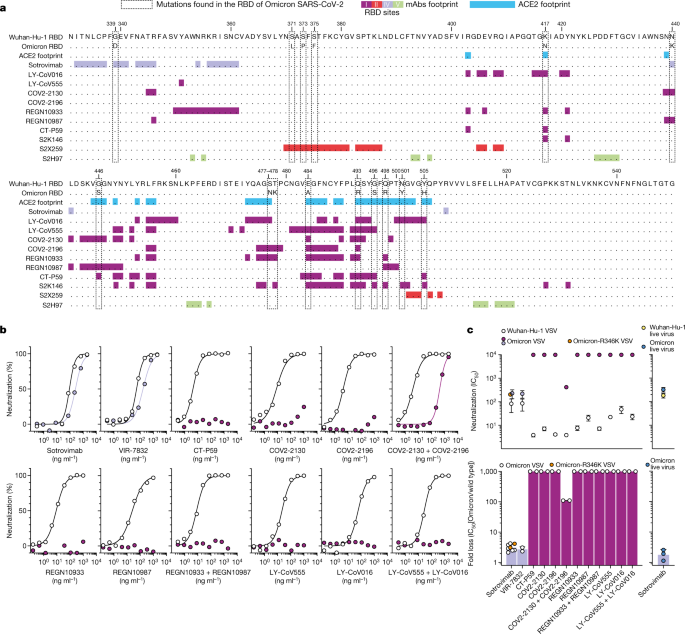
(2021)."). Combining two such ACE2-blocking monoclonal antibodies can provide greater resistance to variant viruses that carry RBM mutations[31](/articles/s41586-021-04386-2#ref-CR31 "Baum, A. et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 369, 1014–1018 (2020)."). The second class of monoclonal antibodies, represented by sotrovimab, do not block ACE2 binding but neutralize SARS-CoV-2 by targeting non-RBM epitopes that are shared across many sarbecoviruses, including SARS-CoV[4](/articles/s41586-021-04386-2#ref-CR4 "Starr, T. N. et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature 597, 97–102 (2021)."),[40](/articles/s41586-021-04386-2#ref-CR40 "Lempp, F. A. et al. Lectins enhance SARS-CoV-2 infection and influence neutralizing antibodies. Nature 598, 342–347 (2021).").Fig. 3: Neutralization of Omicron SARS-CoV-2 VSV pseudovirus by clinical-stage monoclonal antibodies.
a, RBD sequence of SARS-CoV-2 Wuhan-Hu-1 with highlighted footprints of ACE2 (light blue) and monoclonal antibodies (mAbs; coloured according to the RBD antigenic site recognized). The Omicron RBD is also shown, and amino acid substitutions are boxed. b, Neutralization of SARS-CoV-2 VSV pseudoviruses displaying Wuhan-Hu-1 (white) or Omicron (coloured as in Fig. 4b) spike proteins by clinical-stage monoclonal antibodies. Data are representative of at least two independent experiments. Shown is the mean of two technical replicates. c, Geometric mean half-maximum inhibitory concentration (IC50) values for Omicron (coloured as in Fig. 4b) and Wuhan-Hu-1 (white) (top), and geometric mean fold change (bottom). Vero E6 cells were used as target cells. Shown in blue (right) is neutralization of authentic virus by sotrovimab (WA1/2020 versus hCoV-19/USA/WI-WSLH-221686/2021). Non-neutralizing IC50 titres and fold change were set to 104 and 103, respectively. Orange dots for sotrovimab indicate neutralization of Omicron VSV pseudovirus carrying R346K (Omicron-R346K). Data are representative of n = 2 biologically independent experiments for most monoclonal antibodies; for sotrovimab against Omicron VSV n = 6 and for Omicron authentic virus n = 3.
We compared the in vitro neutralizing activity of these therapeutic monoclonal antibodies side-by-side against Wuhan-Hu-1 and Omicron spike proteins using VSV pseudoviruses (Fig. 3). Although sotrovimab had a threefold-reduced potency against Omicron and Omicron-R346K variant VSV pseudoviruses, all of the other (RBM-specific) monoclonal antibodies completely lost their neutralizing activity—with the exception of the combination of COV2-2130 and COV2-2196, for which we determined an approximately 100-fold-reduced potency (Fig. 3b, c). Moreover, sotrovimab exhibited a reduction of less than twofold in neutralizing activity against authentic Omicron SARS-CoV-2 as compared to the WA1/2020 D614G virus (Fig. 3c, Extended Data Fig. 7), consistent with recent reports on S309, the parent of sotrovimab[41](/articles/s41586-021-04386-2#ref-CR41 "VanBlargan, L. A. et al. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat. Med. https://doi.org/10.1038/s41591-021-01678-y
(2022)."),[42](/articles/s41586-021-04386-2#ref-CR42 "Aggarwal, A. et al. SARS-CoV-2 Omicron: evasion of potent humoral responses and resistance to clinical immunotherapeutics relative to viral variants of concern. Preprint at
https://doi.org/10.1101/2021.12.14.21267772
(2021)."). The threefold and less-than-twofold decrease in the neutralizing activity of sotrovimab against pseudoviruses and authentic virus, respectively, is within the currently defined threshold of ‘no change’ as defined by the US Food and Drug Administration (FDA; the FDA fact sheet for sotrovimab denotes no change as a reduction of less than fivefold in susceptibility[43](/articles/s41586-021-04386-2#ref-CR43 "FDA. Fact sheet for healthcare providers: emergency use authorization (EUA) of sotrovimab.
https://www.fda.gov/media/149534/download
(2021).")). Overall, our findings agree with two preliminary reports[16](/articles/s41586-021-04386-2#ref-CR16 "Wilhelm, A. et al. Reduced neutralization of SARS-CoV-2 Omicron variant by vaccine sera and monoclonal antibodies. Preprint at
https://doi.org/10.1101/2021.12.07.21267432
(2021)."),[18](/articles/s41586-021-04386-2#ref-CR18 "Cao, Y. et al. B.1.1.529 escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature
https://doi.org/10.1038/s41586-021-04385-3
(2021).") and, together with serological data, support the conclusion that the Omicron VOC has undergone antigenic shift.We next tested a larger panel of 36 neutralizing NTD- or RBD-specific monoclonal antibodies for which the epitopes have been characterized structurally or assigned to a given antigenic site through competition studies3,4,9,12,44,45 (Fig. 4a, Extended Data Table 2, Extended Data Fig. 8). The four NTD-specific antibodies completely lost activity against Omicron, consistent with the presence of mutations and deletions in the NTD antigenic supersite21,45,46. Three out of the twenty-two monoclonal antibodies that target the RBD antigenic site I (RBM) retained potent neutralizing activity against Omicron, including S2K146, which binds the RBD of SARS-CoV-2, SARS-CoV and other sarbecoviruses through ACE2 molecular mimicry[1](/articles/s41586-021-04386-2#ref-CR1 "Park, Y. J. et al. Antibody-mediated broad sarbecovirus neutralization through ACE2 molecular mimicry. Science https://doi.org/10.1126/science.abm8143
(2022)."). Of the nine monoclonal antibodies that are specific for the conserved RBD site II (refs. [4](/articles/s41586-021-04386-2#ref-CR4 "Starr, T. N. et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature 597, 97–102 (2021)."),[12](/articles/s41586-021-04386-2#ref-CR12 "Piccoli, L. et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell 183, 1024–1042 (2020).")), only S2X259[3](/articles/s41586-021-04386-2#ref-CR3 "Tortorici, M. A. et al. Broad sarbecovirus neutralization by a human monoclonal antibody. Nature 597, 103–108 (2021).") retained activity against Omicron, whereas neutralization was decreased by more than tenfold or abolished for the remaining antibodies. Finally, the S2H97 monoclonal antibody retained neutralizing activity againstOmicron through recognition of the highly conserved cryptic site V (ref. [4](/articles/s41586-021-04386-2#ref-CR4 "Starr, T. N. et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature 597, 97–102 (2021).")). The panel of 44 monoclonal antibodies tested in this study includes members of each of the four classes of neutralizing monoclonal antibodies, defined by their cognate RBD-binding sites (sites I, II, IV and V)[12](/articles/s41586-021-04386-2#ref-CR12 "Piccoli, L. et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell 183, 1024–1042 (2020)."). Our findings show that a member, or members, of each of the four classes can retain Omicron neutralization: S2K146, S2X324 and S2N28 targeting site I, S2X259 targeting site II, sotrovimab targeting site IV and S2H97 targeting site V (Fig. [4b](/articles/s41586-021-04386-2#Fig4)). Several of these monoclonal antibodies cross-react with and neutralize sarbecoviruses beyond the SARS-CoV-2 clade 1b[1](/articles/s41586-021-04386-2#ref-CR1 "Park, Y. J. et al. Antibody-mediated broad sarbecovirus neutralization through ACE2 molecular mimicry. Science
https://doi.org/10.1126/science.abm8143
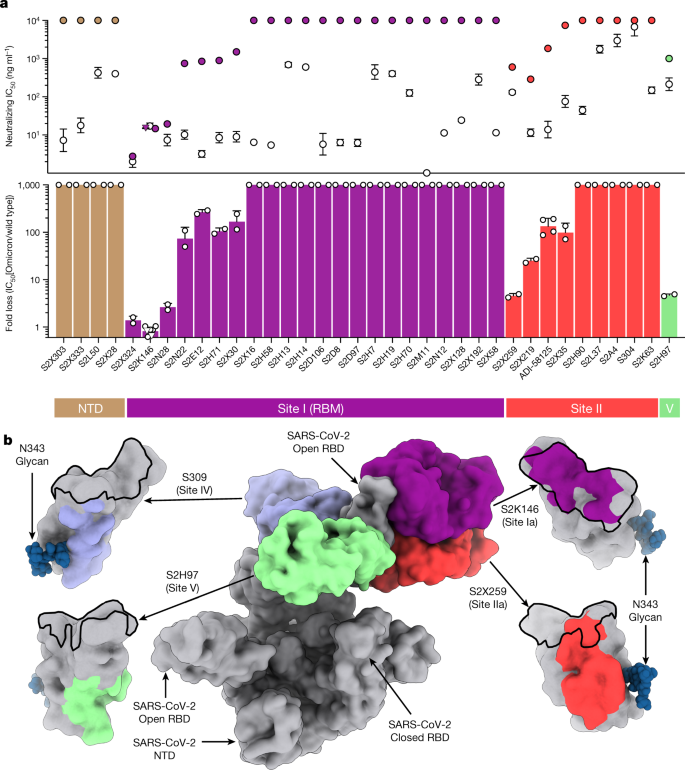
(2022)."),[3](/articles/s41586-021-04386-2#ref-CR3 "Tortorici, M. A. et al. Broad sarbecovirus neutralization by a human monoclonal antibody. Nature 597, 103–108 (2021)."),[4](/articles/s41586-021-04386-2#ref-CR4 "Starr, T. N. et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature 597, 97–102 (2021)."), indicating that targeting of conserved epitopes can lead to neutralization breadth and resilience to antigenic shift associated with viral evolution.Fig. 4: Neutralization of Omicron SARS-CoV-2 VSV pseudovirus by monoclonal antibodies.
a, Top, mean IC50 values for Omicron (coloured as in b) and Wuhan-Hu-1 (white). Bottom, mean fold change for 4 NTD monoclonal antibodies and 32 RBD monoclonal antibodies. Non-neutralizing IC50 titres and fold change were set to 104 and 103, respectively. Triangles for S2K146 indicate neutralization of Omicron carrying R346K. Vero E6 cells were used as target cells. Data are representative of n = 2 biologically independent experiments (except for S2K146, for which n = 6). b, The RBD sites targeted by four monoclonal antibodies that cross-neutralize Omicron are annotated and representative antibodies (the Fv region) bound to spike proteins are shown as a composite. Coloured surfaces on the RBD depict the epitopes and the RBM is shown as a black outline.
Discussion
The high number of substitutions present in the Omicron spike protein marks a pronounced shift in antigenicity and is associated with immune evasion of considerable magnitude for SARS-CoV-2. Antigenic shift of the influenza virus is defined as genetic reassortment of the RNA genome segments, but the mechanism for the abrupt appearance of a large number of mutations in SARS-CoV-2 Omicron spike protein remains to be determined. Although recombination events are a hallmark of coronaviruses47, we and others48 propose that the Omicron shift may result from extensive viral replication in immunodeficient hosts47,49, although we cannot rule out the possibility of a contribution of inter-species ping-pong transmission5 between humans and rodents, as previously described for minks50.
Consistent with the variable decrease in plasma neutralizing antibody titres, we found that only 6 out of a panel of 44 neutralizing monoclonal antibodies retained potent neutralizing activity against Omicron. The monoclonal antibodies that retain neutralization recognize RBD antigenic sites that are conserved in Omicron and other sarbecoviruses. Notably, three of these antibodies bind to the RBM, including one that is a molecular mimic of the ACE2 receptor (S2K146)[1](/articles/s41586-021-04386-2#ref-CR1 "Park, Y. J. et al. Antibody-mediated broad sarbecovirus neutralization through ACE2 molecular mimicry. Science https://doi.org/10.1126/science.abm8143
(2022)."). Collectively, these data may guide future efforts to develop SARS-CoV-2 vaccines and therapies to counteract antigenic shift and future sarbecovirus zoonotic spillovers.Methods
Cell lines
Cell lines used in this study were obtained from ATCC (HEK293T and Vero E6), Thermo Fisher Scientific (Expi-CHO-S cells, FreeStyle 293-F cells and Expi293F cells), Takara (Lenti-X 293T cells) or generated in-house (Vero E6-TMPRSS2)40. Vero-TMPRSS251 cells were cultured at 37 °C in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 10 mM HEPES pH 7.3 and 100 U ml−1 of penicillin–streptomycin and supplemented with 5 µg ml−1 of blasticidin. None of the cell lines used was authenticated. Cell lines were routinely tested for mycoplasma contamination.
Omicron prevalence analysis
The viral sequences and the corresponding metadata were obtained from the GISAID EpiCoV project (https://www.gisaid.org/). Analysis was performed on sequences submitted to GISAID up to 20 December 2021. Spike protein sequences were obtained from the genomic sequences with the exonerate52 2 2.4.0–haf93ef1_3 (https://quay.io/repository/biocontainers/exonerate?tab=tags) using protein to DNA alignment with parameters -m protein2dna –refine full –minintron 999999 –percent 20 and using accession YP_009724390.1 as a reference. Multiple sequence alignment of all human spike proteins was performed with mafft53 7.475–h516909a_0 (https://quay.io/repository/biocontainers/mafft?tab=tags) with parameters –auto –reorder –mapout –keeplength –addfragments using the same reference as above. The –mapout parameter was used to retrieve insertions. Spike sequences that contained more than 10% ambiguous amino acids or that were less than 80% of the canonical protein length were discarded. Figures were generated with R 4.0.2 (https://cran.r-project.org/) using the ggplot2 3.3.2 and sf 0.9-7 packages. To identify each mutation prevalence, missingness (or ambiguous amino acids) was taken into account in both nominator and denominator.
Monoclonal antibodies
Sotrovimab and VIR-7832 (VIR-783254 is derived from sotrovimab, Fc further engineered to carry GAALIE) were produced at WuXi Biologics. Antibody VH and VL sequences for the monoclonal antibodies COV2-2130 (Protein Data Bank (PDB) ID 7L7E), COV2-2196 (PDB ID 7L7E, 7L7D), REGN10933 (PDB ID 6XDG), REGN10987 (PDB ID 6XDG) and ADI-58125 (PCT application WO2021207597, seq. IDs 22301 and 22311) were subcloned into heavy chain (human IgG1) and the corresponding light chain (human Ig κ-chain, Ig λ-chain) expression vectors respectively and produced in transiently transfected ExpiCHO-S cells (Thermo Fisher Scientific, A29133) at 37 °C and 8% CO2. Cells were transfected using ExpiFectamine. Transfected cells were supplemented 1 day after transfection with ExpiCHO Feed and ExpiFectamine CHO Enhancer. Cell culture supernatant was collected eight days after transfection and filtered through a 0.2-µm filter. Recombinant antibodies were affinity purified on an ÄKTA Xpress FPLC device using 5 ml HiTrap MabSelect PrismA columns followed by buffer exchange to histidine buffer (20 mM histidine, 8% sucrose, pH 6) using HiPrep 26/10 desalting columns. Antibody VH and VL sequences for LY-CoV555, LY-CoV016 and CT-P59 were obtained from PDB IDs 7KMG, 7C01 and 7CM4, respectively, and monoclonal antibodies were produced as recombinant IgG1 by ATUM. The remaining monoclonal antibodies were discovered at VIR and have been produced as recombinant IgG1 in ExpiCHO-S cells as described above. The identity of the produced monoclonal antibodies was confirmed by liquid chromatography–mass spectrometry (LC–MS) analysis.
IgG mass quantification by LC–MS intact protein mass analysis
Fc N-linked glycan from monoclonal antibodies was removed by PNGase F after overnight non-denaturing reaction at room temperature. Deglycosylated protein (4 µg) was injected to the LC–MS system to acquire intact MS signal. Thermo MS (Q Exactive Plus Orbitrap) was used to acquire intact protein mass under denaturing condition with m/z window from 1,000 to 6,000. BioPharma Finder 3.2 software was used to deconvolute the raw m/z data to protein average mass. The theoretical mass for each monoclonal antibody was calculated with GPMAW 10.10 software. Post-translational modifications such as N-terminal pyroglutamate cyclization, C-terminal lysine cleavage and formation of 16–18 disulfide bonds were added into the calculation.
Sample donors
Samples were obtained from SARS-CoV-2 convalescent and vaccinated individuals under study protocols approved by the local institutional review boards (Canton Ticino Ethics Committee, Switzerland, Comitato Etico Milano Area 1). All donors provided written informed consent for the use of blood and blood derivatives (such as peripheral blood mononuclear cells, sera or plasma) for research. Plasma samples from convalescent individuals and individuals who had been vaccinated with Ad26.COV2.S, mRNA-1273 or BNT162b2 were obtained from the HAARVI study, approved by the University of Washington Human Subjects Division Institutional Review Board (STUDY00000959). Samples from individuals who had been vaccinated with AZD1222 were obtained from INGM, Ospedale Maggio Policlinico of Milan and approved by the local review board Study Polimmune. Samples from individuals who had been vaccinated with Sputnik V were obtained from healthcare workers at the hospital de Clínicas ‘José de San Martín’, Buenos Aires, Argentina. Samples from individuals who had been vaccinated with Sinopharm were enrolled from Aga Khan University under the institutional review board of the UWARN (United World Antivirus Research Network) study.
Pseudovirus neutralization assays
Generation of VSV pseudovirus used in Vero E6 cells
The plasmid encoding the Omicron SARS-CoV-2 spike variant was generated by overlap PCR mutagenesis of the wild-type plasmid, pcDNA3.1(+)-spike-D1955. Replication defective VSV pseudovirus expressing SARS-CoV-2 spike proteins corresponding to the ancestral Wuhan-Hu-1 virus and the Omicron VOC were generated as previously described46 with some modifications. Lenti-X 293T cells (Takara) were seeded in 15-cm2 dishes at a density of 10 × 106 cells per dish and the following day were transfected with 25 µg of spike expression plasmid with TransIT-Lenti (Mirus, 6600) according to the manufacturer’s instructions. One day after transfection, cells were infected with VSV-luc (VSV-G) with a multiplicity of infection (MOI) of 3 for 1 h, rinsed three times with PBS containing Ca2+ and Mg2+, then incubated for an additional 24 h in complete medium at 37 °C. The cell supernatant was clarified by centrifugation, aliquoted, and frozen at −80 °C.
Generation of VSV pseudovirus used in Vero E6-TMPRSS2 cells
Comparison of Omicron SARS-CoV-2 spike VSV to SARS-CoV-2 G614 spike (YP 009724390.1) VSV and Beta spike VSV used pseudotyped particles prepared as described previously9,56. In brief, HEK293T cells in DMEM supplemented with 10% FBS, 1% penicillin–streptomycin seeded in 10-cm dishes were transfected with the plasmid encoding the corresponding spike glycoprotein using Lipofectamine 2000 (Life Technologies) following the manufacturer’s instructions. One day after transfection, cells were infected with VSV(G*ΔG-luciferase)57 and after 2 h were washed five times with DMEM before adding medium supplemented with anti-VSV-G antibody (I1-mouse hybridoma supernatant, CRL-2700, ATCC). Virus pseudotypes were collected 18–24 h after inoculation, clarified by centrifugation at 2,500_g_ for 5 min, filtered through a 0.45-μm cut-off membrane, concentrated 10 times with a 30-kDa cut-off membrane, aliquoted and stored at −80 °C.
VSV pseudovirus neutralization
Assay performed using Vero E6 cells
Vero E6 cells were grown in DMEM supplemented with 10% FBS and seeded into clear bottom white 96 well plates (PerkinElmer, 6005688) at a density of 20,000 cells per well. The next day, monoclonal antibodies or plasma were serially diluted in pre-warmed complete medium, mixed with pseudoviruses and incubated for 1 h at 37 °C in round bottom polypropylene plates. Medium from cells was aspirated and 50 µl of virus–monoclonal antibody–plasma complexes was added to cells, which were then incubated for 1 h at 37 °C. An additional 100 µl of prewarmed complete medium was then added on top of complexes and cells were incubated for an additional 16–24 h. Conditions were tested in duplicate wells on each plate and eight wells per plate contained untreated infected cells (defining the 0% of neutralization, ‘MAX RLU’ value) and infected cells in the presence of S309 and S2X259 at 20 µg ml−1 each (defining the 100% of neutralization, ‘MIN RLU’ value). Virus–monoclonal antibody–plasma-containing medium was then aspirated from cells and 100 µl of a 1:2 dilution of SteadyLite Plus (PerkinElmer, 6066759) in PBS with Ca2+ and Mg2+ was added to cells. Plates were incubated for 15 min at room temperature and then were analysed on the Synergy-H1 (Biotek). The average relative light units (RLUs) of untreated infected wells (MAX RLUave) was subtracted by the average of MIN RLU (MIN RLUave) and used to normalize percentage of neutralization of individual RLU values of experimental data according to the following formula: (1 − (RLUx – MIN RLUave)/(MAX RLUave – MIN RLUave)) × 100. Data were analysed and visualized with Prism (v.9.1.0). IC50 (monoclonal antibodies) and ID50 (plasma) values were calculated from the interpolated value from the log(inhibitor) versus response, using variable slope (four parameters) nonlinear regression with an upper constraint of ≤100, and a lower constrain equal to 0. Each neutralization experiment was conducted on two independent experiments—that is, biological replicates—in which each biological replicate contains a technical duplicate. IC50 values across biological replicates are presented as arithmetic mean ± s.d. The loss or gain of neutralization potency across spike variants was calculated by dividing the variant IC50/ID50 by the parental IC50/ID50 within each biological replicate, and then visualized as arithmetic mean ± s.d.
Assay performed using Vero E6-TMPRSS2 cells
VeroE6-TMPRSS2 cells were cultured in DMEM with 10% FBS (Hyclone), 1% penicillin–streptomycin and 8 µg ml−1 puromycin (to ensure retention of TMPRSS2) with 5% CO2 in a 37 °C incubator (Thermo Fisher Scientific). Cells were trypsinized using 0.05% trypsin and plated to be at 90% confluence the following day. In an empty half-area 96-well plate, a 1:3 serial dilution of serum was made in DMEM and diluted pseudovirus was then added and incubated at room temperature for 30–60 min before addition of the serum–virus mixture to the cells at 37 °C. Two hours later, 40 μl of a DMEM solution containing 20% FBS and 2% penicillin–streptomycin (Thermo Fisher Scientific, 10,000 units per ml of penicillin and 10,000 µg ml−1 of streptomycin when undiluted) was added to each well. After 17–20 h, 40 μl per well of One-Glo-EX substrate (Promega) was added to the cells and they were incubated in the dark for 5–10 min before reading on a BioTek plate reader. Measurements were done at least in duplicate using distinct batches of pseudoviruses and one representative experiment is shown. RLUs were plotted and normalized in Prism (GraphPad). Nonlinear regression of log(inhibitor) versus normalized response was used to determine IC50 values from curve fits. Normality was tested using the D’Agostino-Pearson test and in the absence of a normal distribution, Kruskal–Wallis tests were used to compare two groups to determine whether differences reached statistical significance. Fold changes were determined by comparing individual IC50 and then averaging the individual fold changes for reporting.
Focus reduction neutralization test
The WA1/2020 strain with a D614G substitution was described previously58. The B.1.1.529 isolate (hCoV-19/USA/WI-WSLH-221686/2021) was obtained from a nasal swab and passaged on Vero-TMPRSS2 cells as described59. The B.1.1.529 isolate was sequenced (GISAID: EPI_ISL_7263803) to confirm the stability of substitutions. All virus experiments were performed in an approved biosafety level 3 (BSL-3) facility.
Serial dilutions of sotrovimab were incubated with 102 focus-forming units of SARS-CoV-2 (WA1/2020 D614G or B.1.1.529) for 1 h at 37 °C. Antibody–virus complexes were added to Vero-TMPRSS2 cell monolayers in 96-well plates and incubated at 37 °C for 1 h. Subsequently, cells were overlaid with 1% (w/v) methylcellulose in MEM. Plates were collected 30 h later (WA1/2020 D614G on Vero-TMPRSS2 cells) or 70 h later (B.1.1.529 on Vero-TMPRSS2 cells) by removal of overlays and fixation with 4% PFA in PBS for 20 min at room temperature. Plates with WA1/2020 D614G were washed and sequentially incubated with an oligoclonal pool of SARS2-2, SARS2-11, SARS2-16, SARS2-31, SARS2-38, SARS2-57 and SARS2-7160 anti-S antibodies. Plates with B.1.1.529 were additionally incubated with a pool of monoclonal antibodies that cross-react with SARS-CoV-1 and bind a CR3022-competing epitope on the RBD61. All plates were subsequently stained with HRP-conjugated goat anti-mouse IgG (Sigma, A8924) in PBS supplemented with 0.1% saponin and 0.1% bovine serum albumin. SARS-CoV-2-infected cell foci were visualized using TrueBlue peroxidase substrate (KPL) and quantitated on an ImmunoSpot microanalyser (Cellular Technologies). Antibody dose response curves were analysed using nonlinear regression analysis with a variable slope (GraphPad Software), and the IC50 was calculated.
VSV pseudovirus entry assays using mouse ACE2
HEK293T (293T) cells (ATCC CRL-11268) were cultured in 10% FBS, 1% penicillin–streptomycin in DMEM at 37 °C in a humidified 8% CO2 incubator. Transient transfection of mouse ACE2 in 293T cells was done 18–24 h before infection using Lipofectamine 2000 (Life Technologies) and an HDM plasmid containing full-length mouse ACE2 (GenBank: Q8R010, synthesized by GenScript) in Opti-MEM. After a 5-h incubation at 37 °C in a humidified 8% CO2 incubator, DMEM with 10% FBS was added and cells were incubated at 37 °C in a humidified 8% CO2 incubator for 18–24 h. Immediately before infection, 293T cells with transient expression of mouse ACE2 were washed with DMEM 1×, then plated with pseudovirus at a 1:75 dilution in DMEM. Infection in DMEM was done with cells between 60% and 80% confluence for 2.5 h before adding FBS and penicillin–streptomycin to final concentrations of 10% and 1%, respectively. After 18–24 h of infection, One-Glo-EX (Promega) was added to the cells and they were incubated in the dark for 5 min before reading on a Synergy H1 Hybrid Multi-Mode plate reader (Biotek). Cell entry levels of pseudovirus generated on different days (biological replicates) were plotted in GraphPad Prism as individual points, and average cell entry across biological replicates was calculated as the geometric mean.
Production of recombinant RBD proteins
SARS-CoV-2 RBD proteins for SPR binding assays (residues 328–531 of the spike protein from GenBank NC_045512.2 with N-terminal signal peptide and C-terminal thrombin cleavage site-TwinStrep-8×His-tag) were expressed in Expi293F (Thermo Fisher Scientific) cells at 37 °C and 8% CO2. Transfections were performed using the ExpiFectamine 293 Transfection Kit (Thermo Fisher Scientific). Cell culture supernatants were collected two to four days after transfection and supplemented with 10× PBS to a final concentration of 2.5× PBS (342.5 mM NaCl, 6.75 mM KCl and 29.75 mM phosphates). SARS-CoV-2 RBDs were purified using cobalt-based immobilized metal affinity chromatography followed by buffer exchange into PBS using a HiPrep 26/10 desalting column (Cytiva) or, for the second batch of Omicron RBD used for SPR, a Superdex 200 Increase 10/300 GL column (Cytiva).
The SARS-CoV-2 Wuhan-Hu-1 and Delta (B.1.617.2) RBD-Avi constructs were synthesized by GenScript into pcDNA3.1- with an N-terminal mu-phosphatase signal peptide and a C-terminal octa-histidine tag, flexible linker and avi tag (GHHHHHHHHGGSSGLNDIFEAQKIEWHE).The boundaries of the construct are N-328RFPN331 and 528KKST531-C (refs. 9,14). Proteins were produced in Expi293F cells (Thermo Fisher Scientific) grown in suspension using Expi293 Expression Medium (Thermo Fisher Scientific) at 37 °C in a humidified 8% CO2 incubator rotating at 130 rpm. Cells grown to a density of 3 million cells per ml were transfected using the the ExpiFectamine 293 Transfection Kit (Thermo Fisher Scientific) and cultivated for 3–5 days. Proteins were purified from clarified supernatants using a nickel HisTrap HP affinity column (Cytiva) and washed with 10 column volumes of 20 mM imidazole, 25 mM sodium phosphate pH 8.0 and 300 mM NaCl before elution on a gradient to 500 mM imidazole. Proteins were biotinylated overnight using the BirA Biotin-Protein Ligase Kit (Avidity) and purified again using theHisTrapHP affinity column. After a wash and elution as before, proteins were buffer-exchanged into 20 mM sodium phosphate pH 8 and 100 mM NaCl, and concentrated using centrifugal filters (Amicon Ultra) before being flash-frozen.
Recombinant production of ACE2 orthologues
Recombinant human ACE2 (residues 19–615 from Uniprot Q9BYF1 with a C-terminal AviTag-10×His-GGG-tag, and N-terminal signal peptide) was produced by ATUM. Protein was purified via Ni Sepharose resin followed by isolation of the monomeric hACE2 by size-exclusion chromatography using a Superdex 200 Increase 10/300 GL column (Cytiva) pre-equilibrated with PBS. The mouse (Mus musculus) ACE2 ectodomain construct (GenBank: Q8R0I0) was synthesized by GenScript and placed into a pCMV plasmid. The domain boundaries for the ectodomain are residues 19–615. The native signal tag was identified using SignalP-5.0 (residues 1–18) and replaced with an N-terminal mu-phosphatase signal peptide. This construct was then fused to a sequence encoding a thrombin cleavage site and a human Fc fragment or an 8×His tag at the C terminus. ACE2-Fc and ACE2-His constructs were produced in Expi293 cells (Thermo Fisher Scientific, A14527) in Gibco Expi293 Expression Medium at 37 °C in a humidified 8% CO2 incubator rotating at 130 rpm. The cultures were transfected using PEI-25K (Polyscience) with cells grown to a density of 3 million cells per ml and cultivated for 4–5 days. Proteins were purified from clarified supernatants using a 1-ml HiTrap Protein A HP affinity column (Cytiva) or a 1-ml HisTrap HP affinity column (Cytiva), concentrated and flash-frozen in 1× PBS, pH 7.4 (10 mM Na2HPO4, 1.8 mM KH2PO4, 2.7 mM KCl, 137 mM NaCl).
ACE2-binding measurements using SPR
Measurements were performed using a Biacore T200 instrument, in triplicate for monomeric human and mouse ACE2 and duplicate for dimeric mouse ACE2. A CM5 chip covalently immobilized with StrepTactin XT (IBA LifeSciences) was used for surface capture of TwinStrepTag-containing RBDs (Wuhan-Hu-1, Alpha, Beta, Omicron, K417N) and a Cytiva Biotin CAPture Kit was used for surface capture of biotinylated RBDs (Delta and Wuhan-Hu-1 used for fold-change comparison to Delta). Two different batches of Omicron RBD were used for the experiments. Running buffer was HBS-EP+ pH 7.4 (Cytiva) and measurements were performed at 25 °C. Experiments were performed with a threefold dilution series of human ACE2 (300, 100, 33, 11 nM) or mouse ACE2 (900, 300, 100, 33 nM) and were run as single-cycle kinetics. Monomeric ACE2-binding data were double-reference-subtracted and fit to a 1:1 binding model using Biacore Evaluation software. High concentrations of dimeric mouse ACE2 exhibited significant binding to the CAP sensor chip reference flow cell.
Statistical analysis
Neutralization measurements were performed in duplicate and RLUs were converted to per cent neutralization and plotted with a nonlinear regression model to determine IC50/ID50 values using GraphPad Prism software (v.9.0.0). Comparisons between two groups of paired two-sided data were made with Wilcoxon rank test.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
Materials generated in this study will be made available on request and may require a material transfer agreement. Access to GISAID (www.gisaid.org) data requires registration. Note: after consulting with the local ethical authority, owing to health and data protection laws relating to the demographic and clinical information contained in the manuscript, we will not be able to fully comply with the requirement to share demographic and clinical data of individual patients and donors in this study.
References
- Park, Y. J. et al. Antibody-mediated broad sarbecovirus neutralization through ACE2 molecular mimicry. Science https://doi.org/10.1126/science.abm8143 (2022).
- Pinto, D. et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295 (2020).
Article ADS CAS PubMed Google Scholar - Tortorici, M. A. et al. Broad sarbecovirus neutralization by a human monoclonal antibody. Nature 597, 103–108 (2021).
Article ADS CAS PubMed Google Scholar - Starr, T. N. et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature 597, 97–102 (2021).
Article ADS CAS PubMed Google Scholar - Telenti, A. et al. After the pandemic: perspectives on the future trajectory of COVID-19. Nature 596, 495–504 (2021).
Article ADS CAS PubMed Google Scholar - Wang, P. et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 593, 130–135 (2021).
Article ADS CAS PubMed Google Scholar - Chen, R. E. et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 27, 717–726 (2021).
Article CAS PubMed PubMed Central Google Scholar - Corti, D., Purcell, L. A., Snell, G. & Veesler, D. Tackling COVID-19 with neutralizing monoclonal antibodies. Cell 184, 3086–3108 (2021).
Article CAS PubMed PubMed Central Google Scholar - McCallum, M. et al. Molecular basis of immune evasion by the Delta and Kappa SARS-CoV-2 variants. Science 374, 1621–1626 (2021).
Article ADS CAS PubMed Google Scholar - Mlcochova, P. et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 599, 114–119 (2021).
Article ADS CAS PubMed PubMed Central Google Scholar - Sheikh, A. et al. SARS-CoV-2 Delta VOC in Scotland: demographics, risk of hospital admission, and vaccine effectiveness. Lancet 397, 2461–2462 (2021).
Article CAS PubMed PubMed Central Google Scholar - Piccoli, L. et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell 183, 1024–1042 (2020).
Article CAS PubMed PubMed Central Google Scholar - Greaney, A. J. et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe 29, 463–476 (2021).
Article CAS PubMed PubMed Central Google Scholar - Walls, A. C. et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181, 281–292 (2020).
Article CAS PubMed PubMed Central Google Scholar - Cele, S. et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature https:///doi.org/10.1038/s41586-021-04387-1 (2021).
- Wilhelm, A. et al. Reduced neutralization of SARS-CoV-2 Omicron variant by vaccine sera and monoclonal antibodies. Preprint at https://doi.org/10.1101/2021.12.07.21267432 (2021).
- Andrews, N. et al. Effectiveness of COVID-19 vaccines against the Omicron variant of concern. Preprint at https://doi.org/10.1101/2021.12.14.21267615 (2021).
- Cao, Y. et al. B.1.1.529 escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature https://doi.org/10.1038/s41586-021-04385-3 (2021).
- Leist, S. R. et al. A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell 183, 1070–1085 (2020).
Article CAS PubMed PubMed Central Google Scholar - Dinnon, K. H. III et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 586, 560–566 (2020).
Article ADS PubMed PubMed Central Google Scholar - Collier, D. A. et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature 593, 136–141 (2021).
Article ADS CAS PubMed Google Scholar - Starr, T. N. et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 182, 1295–1310 (2020).
Article CAS PubMed PubMed Central Google Scholar - Shuai, H. et al. Emerging SARS-CoV-2 variants expand species tropism to murines. EBioMedicine 73, 103643 (2021).
Article CAS PubMed PubMed Central Google Scholar - Pan, T. et al. Infection of wild-type mice by SARS-CoV-2 B.1.351 variant indicates a possible novel cross-species transmission route. Sig. Transduct. Target Ther. 6, 420 (2021).
Article CAS Google Scholar - Hoffmann, M. et al. The Omicron variant is highly resistant against antibody-mediated neutralization—implications for control of the COVID-19 pandemic. Cell https://doi.org/10.1016/j.cell.2021.12.032 (2021).
- Nicholls, J. & Chan Chi-wai, M. HKUMed finds Omicron SARS-CoV-2 can infect faster and better than Delta in human bronchus but with less severe infection in lung. www.med.hku.hk/en/news/press/20211215-omicron-sars-cov-2-infection (2021).
- Bassi, J. et al. Poor neutralization and rapid decay of antibodies to SARS-CoV-2 variants in vaccinated dialysis patients. Preprint at https://doi.org/10.1101/2021.10.05.21264054 (2021).
- Khoury, D. S. et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 27, 1205–1211 (2021).
Article CAS PubMed Google Scholar - Stamatatos, L. et al. mRNA vaccination boosts cross-variant neutralizing antibodies elicited by SARS-CoV-2 infection. Science https://doi.org/10.1126/science.abg9175 (2021).
- Bergstrom, J. J., Xu, H. & Heyman, B. Epitope-specific suppression of IgG responses by passively administered specific IgG: evidence of epitope masking. Front. Immunol. 8, 238 (2017).
Article PubMed PubMed Central Google Scholar - Baum, A. et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 369, 1014–1018 (2020).
Article ADS CAS PubMed Google Scholar - Hansen, J. et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 369, 1010–1014 (2020).
Article ADS CAS PubMed PubMed Central Google Scholar - Shi, R. et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 584, 120–124 (2020).
Article ADS CAS PubMed Google Scholar - Gottlieb, R. L. et al. Effect of bamlanivimab as monotherapy or in combination with etesevimab on viral load in patients with mild to moderate COVID-19: a randomized clinical trial. JAMA 325, 632–644 (2021).
Article CAS PubMed PubMed Central Google Scholar - Jones, B. E. et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med. 13, eabf1906 (2021).
Article CAS PubMed PubMed Central Google Scholar - Dong, J. et al. Genetic and structural basis for SARS-CoV-2 variant neutralization by a two-antibody cocktail. Nat. Microbiol. 6, 1233–1244 (2021).
Article CAS PubMed PubMed Central Google Scholar - Zost, S. J. et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 584, 443–449 (2020).
Article CAS PubMed PubMed Central Google Scholar - Thomson, E. C. et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell 184, 1171–1187 (2021).
Article CAS PubMed PubMed Central Google Scholar - Starr, T. N. et al. ACE2 binding is an ancestral and evolvable trait of sarbecoviruses. Preprint at https://doi.org/10.1101/2021.07.17.452804 (2021).
- Lempp, F. A. et al. Lectins enhance SARS-CoV-2 infection and influence neutralizing antibodies. Nature 598, 342–347 (2021).
Article ADS CAS PubMed Google Scholar - VanBlargan, L. A. et al. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat. Med. https://doi.org/10.1038/s41591-021-01678-y (2022).
- Aggarwal, A. et al. SARS-CoV-2 Omicron: evasion of potent humoral responses and resistance to clinical immunotherapeutics relative to viral variants of concern. Preprint at https://doi.org/10.1101/2021.12.14.21267772 (2021).
- FDA. Fact sheet for healthcare providers: emergency use authorization (EUA) of sotrovimab. https://www.fda.gov/media/149534/download (2021).
- Tortorici, M. A. et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science 370, 950–957 (2020).
Article ADS CAS PubMed PubMed Central Google Scholar - McCallum, M. et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell 184, 2332–2347 (2021).
Article CAS PubMed PubMed Central Google Scholar - McCallum, M. et al. SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science 373, 648–654 (2021).
Article ADS CAS PubMed Google Scholar - Fischer, W. et al. HIV-1 and SARS-CoV-2: patterns in the evolution of two pandemic pathogens. Cell Host Microbe 29, 1093–1110 (2021).
Article CAS PubMed PubMed Central Google Scholar - Kupferschmidt, K. Where did ‘weird’ Omicron come from? Science 374, 1179 (2021).
Article ADS PubMed Google Scholar - Corey, L. et al. SARS-CoV-2 variants in patients with immunosuppression. N. Engl. J. Med. 385, 562–566 (2021).
Article CAS PubMed Google Scholar - Oude Munnink, B. B. et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 371, 172–177 (2021).
Article ADS CAS PubMed Google Scholar - Zang, R. et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 5, eabc3582 (2020).
Article CAS PubMed PubMed Central Google Scholar - Maziarz, R. T. et al. Control of an outbreak of human parainfluenza virus 3 in hematopoietic stem cell transplant recipients. Biol. Blood Marrow Transplant. 16, 192–198 (2010).
Article PubMed Google Scholar - Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Article CAS PubMed PubMed Central Google Scholar - Yamin, R. et al. Fc-engineered antibody therapeutics with improved anti-SARS-CoV-2 efficacy. Nature 599, 465–470 (2021).
Article ADS CAS PubMed Google Scholar - Giroglou, T. et al. Retroviral vectors pseudotyped with severe acute respiratory syndrome coronavirus S protein. J. Virol. 78, 9007–9015 (2004).
Article CAS PubMed PubMed Central Google Scholar - Walls, A. C. et al. Elicitation of broadly protective sarbecovirus immunity by receptor-binding domain nanoparticle vaccines. Cell 184, 5432–5447 (2021).
Article CAS PubMed PubMed Central Google Scholar - Kaname, Y. et al. Acquisition of complement resistance through incorporation of CD55/decay-accelerating factor into viral particles bearing baculovirus GP64. J. Virol. 84, 3210–3219 (2010).
Article CAS PubMed PubMed Central Google Scholar - Plante, J. A. et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 592, 116–121 (2021).
Article ADS CAS PubMed Google Scholar - Imai, M. et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl Acad. Sci. USA 117, 16587–16595 (2020).
Article CAS PubMed PubMed Central Google Scholar - Liu, Z. et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe 29, 477–488 (2021).
Article CAS PubMed PubMed Central Google Scholar - VanBlargan, L. A. et al. A potently neutralizing SARS-CoV-2 antibody inhibits variants of concern by utilizing unique binding residues in a highly conserved epitope. Immunity 54, 2399–2416 (2021).
Article CAS PubMed PubMed Central Google Scholar - Cathcart, A. L. et al. The dual function monoclonal antibodies VIR-7831 and VIR-7832 demonstrate potent in vitro and in vivo activity against SARS-CoV-2. Preprint at https://doi.org/10.1101/2021.03.09.434607 (2021).
- Gupta, A. et al. Early treatment for Covid-19 with SARS-CoV-2 neutralizing antibody sotrovimab. N. Engl. J. Med. 385, 1941–1950 (2021).
Article CAS PubMed Google Scholar - Kim, C. et al. A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein. Nat. Commun. 12, 288 (2021).
Article CAS PubMed PubMed Central Google Scholar - Ryu, D. K. et al. Therapeutic effect of CT-P59 against SARS-CoV-2 South African variant. Biochem. Biophys. Res. Commun. 566, 135–140 (2021).
Article CAS PubMed PubMed Central Google Scholar - Baum, A. et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science 370, 1110–1115 (2020).
Article ADS CAS PubMed PubMed Central Google Scholar - Copin, R. et al. The monoclonal antibody combination REGEN-COV protects against SARS-CoV-2 mutational escape in preclinical and human studies. Cell 184, 3949–3961 (2021).
Article CAS PubMed PubMed Central Google Scholar - Weinreich, D. M. et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N. Engl. J. Med. 384, 238–251 (2021).
Article CAS PubMed Google Scholar - Chen, P. et al. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with Covid-19. N. Engl. J. Med. 384, 229–237 (2021).
Article CAS PubMed Google Scholar - Group, A.-T. L.-C. S. et al. A neutralizing monoclonal antibody for hospitalized patients with Covid-19. N. Engl. J. Med. 384, 905–914 (2021).
Article Google Scholar - Belk, J., Deveau, L. M., Rappazzo, C. G., Walker, L. & Wec, A. PCT application WO/2021/207597: Compounds specific to coronavirus S protein and uses thereof. (Adagio Therapeutics, 2021).
Acknowledgements
We thank H. Tani for providing the reagents necessary for preparing VSV pseudotyped viruses. This study was supported by the National Institute of Allergy and Infectious Diseases (DP1AI158186 and HHSN272201700059C to D.V.), a Pew Biomedical Scholars Award (D.V.), an Investigators in the Pathogenesis of Infectious Disease Awards from the Burroughs Wellcome Fund (D.V.), Fast Grants (D.V.) and the National Institute of General Medical Sciences (5T32GM008268-32 to S.K.Z.). D.V. is an Investigator of the Howard Hughes Medical Institute. O.G. is funded by the Swiss Kidney Foundation. This work was supported, in part, by the National Institutes of Allergy and Infectious Diseases Center for Research on Influenza Pathogenesis (HHSN272201400008C), the Center for Research on Influenza Pathogenesis and Transmission (CRIPT) (75N93021C00014) and the Japan Program for Infectious Diseases Research and Infrastructure (JP21wm0125002) from the Japan Agency for Medical Research and Development (AMED). This work was supported in part through National Institutes of Health USA grant U01 AI151698 for the United World Antivirus Research Network (UWARN).
Author information
Author notes
- These authors contributed equally: Elisabetta Cameroni, John E. Bowen, Laura E. Rosen, Christian Saliba
- These authors jointly supervised this work: Herbert W. Virgin, Antonio Lanzavecchia, Gyorgy Snell, David Veesler, Davide Corti
Authors and Affiliations
- Humabs Biomed SA, a subsidiary of Vir Biotechnology, Bellinzona, Switzerland
Elisabetta Cameroni, Christian Saliba, Katja Culap, Dora Pinto, Anna De Marco, Fabrizia Zatta, Jessica Bassi, Chiara Silacci-Fregni, Luca Piccoli, Matteo Samuele Pizzuto, Antonio Lanzavecchia & Davide Corti - Department of Biochemistry, University of Washington, Seattle, WA, USA
John E. Bowen, Samantha K. Zepeda, Kaitlin R. Sprouse, Alexandra C. Walls & David Veesler - Vir Biotechnology, San Francisco, CA, USA
Laura E. Rosen, Julia di Iulio, Hannah Kaiser, Julia Noack, Nisar Farhat, Nadine Czudnochowski, Colin Havenar-Daughton, Josh R. Dillen, Abigail E. Powell, Alex Chen, Cyrus Maher, Li Yin, David Sun, Leah Soriaga, Christy Hebner, Lisa A. Purcell, Amalio Telenti, Herbert W. Virgin & Gyorgy Snell - Department of Medicine, Washington University of School of Medicine, St Louis, MO, USA
Laura A. VanBlargan & Michael S. Diamond - ATUM, Newark, CA, USA
Claes Gustafsson - Division of Allergy and Infectious Diseases, University of Washington, Seattle, WA, USA
Nicholas M. Franko, Jenni Logue & Helen Chu - Department of Paediatrics and Child Health, Aga Khan University, Karachi, Pakistan
Najeeha Talat Iqbal - Instituto de Investigaciones Biomédicas en Retrovirus y SIDA (INBIRS), Facultad de Medicina, Universidad de Buenos Aires, Buenos Aires, Argentina
Ignacio Mazzitelli & Jorge Geffner - National Institute of Molecular Genetics, Milan, Italy
Renata Grifantini & Antonio Lanzavecchia - Infectious Disease Unit, Fondazione IRCCS Ca’ Granda, Ospedale Maggiore Policlinico, Milan, Italy
Andrea Gori - Department of Biomedical and Clinical Sciences ‘L.Sacco’ (DIBIC), Università di Milano, Milan, Italy
Agostino Riva - Faculty of Biomedical Sciences, Università della Svizzera italiana, Lugano, Switzerland
Olivier Giannini, Alessandro Ceschi & Paolo Ferrari - Department of Medicine, Ente Ospedaliero Cantonale, Bellinzona, Switzerland
Olivier Giannini & Pietro E. Cippà - Clinical Trial Unit, Ente Ospedaliero Cantonale, Lugano, Switzerland
Alessandro Ceschi - Division of Clinical Pharmacology and Toxicology, Institute of Pharmacological Sciences of Southern Switzerland, Ente Ospedaliero Cantonale, Lugano, Switzerland
Alessandro Ceschi - Department of Clinical Pharmacology and Toxicology, University Hospital Zurich, Zurich, Switzerland
Alessandro Ceschi - Division of Nephrology, Ente Ospedaliero Cantonale, Lugano, Switzerland
Paolo Ferrari & Pietro E. Cippà - Clinical School, University of New South Wales, Sydney, New South Wales, Australia
Paolo Ferrari - Faculty of Medicine, University of Zurich, Zurich, Switzerland
Pietro E. Cippà - Clinical Research Unit, Clinica Luganese Moncucco, Lugano, Switzerland
Alessandra Franzetti-Pellanda - Clinic of Internal Medicine and Infectious Diseases, Clinica Luganese Moncucco, Lugano, Switzerland
Christian Garzoni - Influenza Research Institute, Department of Pathobiological Sciences, School of Veterinary Medicine, University of Wisconsin–Madison, Madison, WI, USA
Peter J. Halfmann & Yoshihiro Kawaoka - Division of Virology, Department of Microbiology and Immunology, Institute of Medical Science, University of Tokyo, Tokyo, Japan
Yoshihiro Kawaoka - The Research Center for Global Viral Diseases, National Center for Global Health and Medicine Research Institute, Tokyo, Japan
Yoshihiro Kawaoka - Howard Hughes Medical Institute, Seattle, WA, USA
Alexandra C. Walls & David Veesler - Department of Pathology and Immunology, Washington University School of Medicine, St Louis, MO, USA
Michael S. Diamond & Herbert W. Virgin - Department of Molecular Microbiology, Washington University School of Medicine, St Louis, MO, USA
Michael S. Diamond - Department of Internal Medicine, UT Southwestern Medical Center, Dallas, TX, USA
Herbert W. Virgin
Authors
- Elisabetta Cameroni
You can also search for this author inPubMed Google Scholar - John E. Bowen
You can also search for this author inPubMed Google Scholar - Laura E. Rosen
You can also search for this author inPubMed Google Scholar - Christian Saliba
You can also search for this author inPubMed Google Scholar - Samantha K. Zepeda
You can also search for this author inPubMed Google Scholar - Katja Culap
You can also search for this author inPubMed Google Scholar - Dora Pinto
You can also search for this author inPubMed Google Scholar - Laura A. VanBlargan
You can also search for this author inPubMed Google Scholar - Anna De Marco
You can also search for this author inPubMed Google Scholar - Julia di Iulio
You can also search for this author inPubMed Google Scholar - Fabrizia Zatta
You can also search for this author inPubMed Google Scholar - Hannah Kaiser
You can also search for this author inPubMed Google Scholar - Julia Noack
You can also search for this author inPubMed Google Scholar - Nisar Farhat
You can also search for this author inPubMed Google Scholar - Nadine Czudnochowski
You can also search for this author inPubMed Google Scholar - Colin Havenar-Daughton
You can also search for this author inPubMed Google Scholar - Kaitlin R. Sprouse
You can also search for this author inPubMed Google Scholar - Josh R. Dillen
You can also search for this author inPubMed Google Scholar - Abigail E. Powell
You can also search for this author inPubMed Google Scholar - Alex Chen
You can also search for this author inPubMed Google Scholar - Cyrus Maher
You can also search for this author inPubMed Google Scholar - Li Yin
You can also search for this author inPubMed Google Scholar - David Sun
You can also search for this author inPubMed Google Scholar - Leah Soriaga
You can also search for this author inPubMed Google Scholar - Jessica Bassi
You can also search for this author inPubMed Google Scholar - Chiara Silacci-Fregni
You can also search for this author inPubMed Google Scholar - Claes Gustafsson
You can also search for this author inPubMed Google Scholar - Nicholas M. Franko
You can also search for this author inPubMed Google Scholar - Jenni Logue
You can also search for this author inPubMed Google Scholar - Najeeha Talat Iqbal
You can also search for this author inPubMed Google Scholar - Ignacio Mazzitelli
You can also search for this author inPubMed Google Scholar - Jorge Geffner
You can also search for this author inPubMed Google Scholar - Renata Grifantini
You can also search for this author inPubMed Google Scholar - Helen Chu
You can also search for this author inPubMed Google Scholar - Andrea Gori
You can also search for this author inPubMed Google Scholar - Agostino Riva
You can also search for this author inPubMed Google Scholar - Olivier Giannini
You can also search for this author inPubMed Google Scholar - Alessandro Ceschi
You can also search for this author inPubMed Google Scholar - Paolo Ferrari
You can also search for this author inPubMed Google Scholar - Pietro E. Cippà
You can also search for this author inPubMed Google Scholar - Alessandra Franzetti-Pellanda
You can also search for this author inPubMed Google Scholar - Christian Garzoni
You can also search for this author inPubMed Google Scholar - Peter J. Halfmann
You can also search for this author inPubMed Google Scholar - Yoshihiro Kawaoka
You can also search for this author inPubMed Google Scholar - Christy Hebner
You can also search for this author inPubMed Google Scholar - Lisa A. Purcell
You can also search for this author inPubMed Google Scholar - Luca Piccoli
You can also search for this author inPubMed Google Scholar - Matteo Samuele Pizzuto
You can also search for this author inPubMed Google Scholar - Alexandra C. Walls
You can also search for this author inPubMed Google Scholar - Michael S. Diamond
You can also search for this author inPubMed Google Scholar - Amalio Telenti
You can also search for this author inPubMed Google Scholar - Herbert W. Virgin
You can also search for this author inPubMed Google Scholar - Antonio Lanzavecchia
You can also search for this author inPubMed Google Scholar - Gyorgy Snell
You can also search for this author inPubMed Google Scholar - David Veesler
You can also search for this author inPubMed Google Scholar - Davide Corti
You can also search for this author inPubMed Google Scholar
Contributions
D.C., G.S., M.S.P., L.P. and D.V. conceived the research and designed the study. D.C., D.P., E.C., L.E.R., G.S., M.S.P., L.P., J.E.B., S.K.Z., A.C.W. and D.V. designed experiments. E.C. and K.C. designed and performed mutagenesis for the spike mutant expression plasmids. C.S., D.P., H.K., J.N., N.F., K.R.S. and S.K.Z. produced pseudoviruses. C.S., J.E.B., S.K.Z., D.P., F.Z., J.B., C.S.-F. and A.D.M. performed pseudovirus entry or neutralization assays. C.S., K.C. and E.C. expressed antibodies. L.A.V., P.J.H. and Y.K. isolated and propagated the SARS-CoV-2 Omicron live virus. L.A.V. performed authentic virus neutralization assays. M.S.D. supervised the research on authentic virus neutralization assays. L.E.R. performed binding assays. C. Gustafsson, S.K.Z., A.C.W., N.C., A.E.P. and J.R.D. synthesized expression plasmids and expressed and purified ACE2 and RBD proteins. C.S. and A.C. produced and performed quality control of monoclonal antibodies. J.d.I., C.M., L.Y., D.S. and L.S. performed bioinformatic and epidemiology analyses. C.S., D.P., L.P., L.E.R., M.S.P. and A.D.M. interpreted data. E.C., C.S., F.Z., A.D.M., K.C., D.P., J.E.B., L.E.R., S.K.Z., A.C.W., D.V., A.T., G.S. and D.C. analysed data. A.R., O.G., C. Garzoni, A.C., P.F., A.F.-P., H.C., N.M.F., J.L., N.T.I., I.M., J.G., R.G., A.G, P.E.C. and C.H.-D. contributed to the recruitment of donors and collection of plasma samples. D.C., A.L., H.W.V., G.S., A.T., C.H., L.A.P. and D.V. wrote the manuscript with input from all authors.
Corresponding authors
Correspondence toGyorgy Snell, David Veesler or Davide Corti.
Ethics declarations
Competing interests
E.C., K.C., C.S., D.P., F.Z., A.D.M., A.L., L.P., M.S.P., D.C., H.K., J.N., N.F., J.d.I., L.E.R., N.C., C.H.-D., K.R.S., J.R.D., A.E.P., A.C., C.M., L.Y., D.S., L.S., L.A.P., C.H., A.T., H.W.V. and G.S. are employees of Vir Biotechnology and may hold shares in Vir Biotechnology. L.A.P. is a former employee and shareholder in Regeneron Pharmaceuticals. Regeneron provided no funding for this work. H.W.V. is a founder and holds shares in PierianDx and Casma Therapeutics. Neither company provided resources. The D.V. laboratory has received a sponsored research agreement from Vir Biotechnology. H.C. reported consulting with Ellume, Pfizer, the Bill and Melinda Gates Foundation, Glaxo Smith Kline and Merck. She has received research funding from Emergent Ventures, Gates Ventures, Sanofi Pasteur and the Bill and Melinda Gates Foundation, and support and reagents from Ellume and Cepheid outside of the submitted work. M.S.D. is a consultant for Inbios, Vir Biotechnology, Senda Biosciences and Carnival Corporation, and is on the Scientific Advisory Boards of Moderna and Immunome. The M.S.D. laboratory has received funding support in sponsored research agreements from Moderna, Vir Biotechnology and Emergent BioSolutions. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Peer review
Peer review information
Nature thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 2 Amino acid substitutions and their prevalence in the Omicron RBD.
a, SARS-CoV-2 spike protein in fully open conformation (PDB: 7K4N) with positions of mutated residues in Omicron highlighted on one protomer in green or red spheres in or outside the ACE2 footprint (ACE2), respectively. RBM is defined by a 6 Å cut-off in the RBD-ACE2 interface38. Not all Omicron mutations are shown. b, Substitutions and their prevalence in Omicron sequences reported in GISAID as of 20 December 2021 (ambiguous amino acid substitutions are indicated with strikethrough cells). Shown are also the prevalence in Omicron (%) and substitutions found in other variants. K417N mutation in Delta is found only in a fraction of sequences.
Extended Data Fig. 3 Amino acid substitutions and their prevalence in the Omicron NTD.
Sequences reported in GISAID as of 20 December 2021 (ambiguous amino acid substitutions are marked with strikethrough cells). Shown are also the substitutions found in other variants.
Extended Data Fig. 4 Amino acid substitutions and their prevalence in Omicron S2.
Sequences reported in GISAID as of 20 December 2021 (ambiguous amino acid substitutions are marked with strikethrough cells). Shown are also the substitutions found in other variants.
Extended Data Fig. 5 Characteristics of emergent mutations of Omicron.
a, Shared mutations of Omicron with other sarbecovirus and with VOC. b, Since the beginning of the pandemic there is a progressive coalescence of Omicron-defining mutations into non-Omicron haplotypes that may carry as many as 10 of the Omicron-defining mutations. c, Pango lineages (dots) rarely carry more than 10-15 lineage-defining mutations. d, Exceptionally, some non-Omicron haplotypes may carry up to a maximum 19 Omicron-defining mutations. Shown are selected exceptional haplotypes. Spike G142D and Y145del may also be noted as G142del and Y145D.
Extended Data Fig. 6 SPR analysis of human and mouse ACE2.
a, Full fit results for one representative replicate from each quantifiable SPR dataset with a monomeric analyte (1:1 binding model). b, Single-cycle kinetics SPR analysis of dimeric mouse ACE2 binding to six RBD variants. Dimeric ACE2 is injected successively at 33, 100, 300, and 900 nM. White and grey stripes indicate association and dissociation phases, respectively. The asterisk indicates where high concentrations of dimeric mouse ACE2 are non-specifically binding to the sensor chip surface (Delta experiment was performed separately from the other RBD variants, with a different capture tag and chip surface).
Extended Data Fig. 7 Neutralization of SARS-CoV-2 Omicron live virus by sotrovimab in Vero-TMPRSS2 cells.
a–f, Neutralization curves in Vero-TMPRSS2 cells comparing the sensitivity of SARS-CoV-2 strains with sotrovimab with WA1/2020 D614G and hCoV-19/USA/WI-WSLH-221686/2021 (an infectious clinical isolate of Omicron from a symptomatic individual in the United States). Shown are three independent experiments performed in technical duplicate.
Extended Data Fig. 8 Neutralization of WT (D614) and Omicron SARS-CoV-2 Spike pseudotyped virus by a panel of 36 monoclonal antibodies.
a–c, Neutralization of SARS-CoV-2 VSV pseudoviruses carrying wild-type D614 (grey) or Omicron (orange) spike protein by NTD-targeting (a) and RBD-targeting (b, c) monoclonal antibodies (b, site I; c, sites II and V). Data are representative of two independent experiments. Shown is the mean of 2 technical replicates.
Extended Data Table 1 Demographics of enrolled donors
Extended Data Table 2 Properties of tested monoclonal antibodies
Supplementary information
Supplementary Information
This Supplementary Information contains Supplementary Figs. 1–3, which show neutralization plots of individual plasma donor shown in Fig. 2, and Supplementary Table 1, which shows the effect of point mutations in RBD on its expression and binding to human ACE2 as measured by flow cytometry (FACS) and surface plasmon resonance (SPR).
Reporting Summary
Rights and permissions
About this article
Cite this article
Cameroni, E., Bowen, J.E., Rosen, L.E. et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift.Nature 602, 664–670 (2022). https://doi.org/10.1038/s41586-021-04386-2
- Received: 12 December 2021
- Accepted: 23 December 2021
- Published: 23 December 2021
- Issue Date: 24 February 2022
- DOI: https://doi.org/10.1038/s41586-021-04386-2