N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications (original) (raw)
Main
Post-transcriptional modification of mRNA has emerged as an important mechanism for gene regulation. Among internal mRNA modifications, m6A is by far the most abundant, tagging over 10,000 mRNAs and long noncoding RNAs1,2. It is a reversible modification, and both methyltranferases and demethylases have been reported. In 2014, we and others reported that Mettl3 and Mettl14 formed a heterodimer and served as core components of m6A methyltransferase3,4. Both Mettl3 and Mettl14 are required for m6A formation: in the heterodimer, Mettl3 is the enzymatic subunit and Mettl14 is required for RNA substrate recognition and maintenance of proper Mettl3 conformation5,6,7.
The functional role of m6A in gene expression regulation has been studied extensively8, but its importance in development at organismal levels remains largely unknown. Recently, two studies reported that children born with homozygous missense mutations in the gene FTO (fat mass and obesity-associated protein), which encodes an m6A demethylase9, display severe neurodevelopmental disorders, including microcephaly, functional brain deficits and psychomotor delay, suggesting an essential, yet unexplored, role of m6A RNA modification in brain development10,11. To investigate potential m6A function in early neuronal development, we deleted Mettl14 in mouse embryonic NSCs, as self-renewing and multipotent NSCs give rise to the entire brain, and defects in NSC activities have been shown to underlie various neurodevelopmental disorders12. In vitro, NSCs lacking Mettl14 displayed robust decreases in proliferation accompanied by premature differentiation, suggesting that m6A is required for NSC self-renewal. Consistent with this, in vivo analysis in _Mettl14-_null embryos revealed that NSCs, which are also known as radial glial cells (RGCs), in the ventricular zone (VZ) showed a decrease in number relative to those seen in control mice, and this reduction was accompanied by fewer late-born cortical neurons. Mechanistically, we observed a genome-wide increase in specific histone modifications in _Mettl14_-knockout (KO) NSCs. Notably, gene-by-gene analysis suggested that those changes were correlated with changes in gene expression and observed developmental phenotypes, suggesting that m6A-regulated histone modification underlies alterations in NSC gene expression and activity. Finally, we present evidence that m6A regulation of histone modification alters the stability of mRNAs encoding histone modifiers. Overall, our results show, for the first time, a key role for mRNA modification in NSCs and brain development.
Results
Mettl14 knockout decreases NSC proliferation and promotes premature NSC differentiation in vitro
To assess Mettl14 loss of function in vivo, we generated _Mettl14_–conditional knockout mice (Mettl14 f/f) by flanking Mettl14 exon 2 with loxP sites. Cre-mediated exon 2 excision results in an out-of-frame mutation that abolishes Mettl14 function (Supplementary Fig. 1a,b). To assess whether the KO strategy deletes Mettl14 in vivo, we evaluated whether Mettl14 was deleted globally using EIIa-cre transgenic mice, which express Cre at zygotic stages (Supplementary Fig. 1c,d). Mettl14 +/− heterozygotes were viable and fertile and exhibited no discernible morphological or growth abnormalities, whereas no Mettl14 −/− offspring were observed after crosses of Mettl14 +/− mice (Supplementary Table 1). We then collected embryos resulting from crosses of heterozygotes at embryonic day 7.5 (E7.5), E8.5 and E9.5 for genotyping. Mettl14 −/− embryos were identified at Mendelian ratios when we combined genotyping results from all three stages (Supplementary Table 2). But most Mettl14 −/− embryos were dead and many had regressed (Supplementary Fig. 1e), indicating that Mettl14 activity is required for early embryogenesis, a phenotype similar to that of global _Mettl3_-KO mice13. Of seven Mettl14 −/− embryos identified at either E7.5 or E8.5, four were male and three were female, suggesting that phenotypes were not gender specific (Supplementary Fig. 1f).
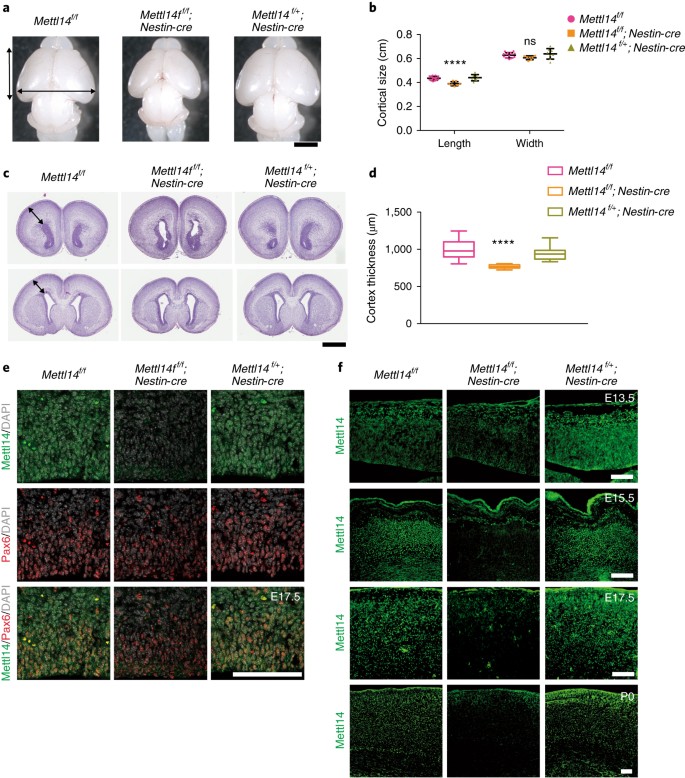
We then assessed the potential effects of Mettl14 deletion in NSCs. To do so, we crossed Mettl14 f/f mice with a Nestin-Cre transgenic line to generate Mettl14 f/f ;Nestin-Cre (_Mettl14_-cKO) mice and littermate controls, including Mettl14 f/+ ;Nestin-cre(heterozygous) and Mettl14 f/f (non-deleted) mice. Newborn pups were alive and showed no overt morphologic phenotypes (Supplementary Fig. 1g) and normal body weight (Supplementary Fig. 1h). However, all _Mettl14_-cKO mice were dead within the first neonatal week (Supplementary Fig. 1i). When we examined the brains of postnatal day 0 (P0) _Mettl14_-cKO pups, we observed no anomalies in gross anatomy, but we found moderately reduced cortical length (Fig. 1a,b). Hematoxylin and eosin (H&E) staining of coronal sections of P0 mouse brain revealed enlargement of the ventricle and a 23% decrease in cortical thickness in _Mettl14_-cKO brains relative to littermate Mettl14 f/f controls (Fig. 1c,d). We next examined Mettl14 expression in RGCs by carrying out Mettl14 and Pax6 co-immunostaining on coronal sections of E17.5 brain from nondeleted, cKO and heterozygous mice. Mettl14 was readily detectible in Pax6+ cells in the cortex of nondeleted and heterozygous controls, but not in cKO mice (Fig. 1e). Together, these results suggest that Mettl14 is required for normal function of NSCs that serve as cortical progenitors.
Fig. 1: Mettl14 regulates the size of mouse cerebral cortex.
a, Representative images of whole brains from Mettl14 f/f (wild-type (WT); left), Mettl14 f/f ;Nestin-cre (KO; middle) and Mettl14 f/+ ;Nestin-cre (heterozygous (Het); right) mice pups at P0; black arrows indicate cortex length and width. Scale bar represents 2 mm. b, Quantification of cortical length and width at P0; one-way ANOVA (WT: n = 16; KO: n = 7,; Het: n = 8 P0 brains; length, P = 2.559 × 10–5, F(2, 28) = 15.79; width, P = 0.0869, F (2, 28) = 2.669) followed by Bonferroni’s post hoc test (length, WT versus KO, P = 4.358 × 10–5, 95% confidence interval (C.I.) = 0.02383–0.06534, WT versus Het, P = 0.9999, 95% C.I. = –0.02521–0.01446; width, WT versus KO, P = 0.2141, 95% C.I. = –0.008986–0.05154, WT versus Het, P = 0.6633, 95% C.I. = –0.04098–0.01686). c, Representative images of coronal sections of P0 brains stained with H&E; black arrows indicate cortical thickness. Section shown in upper panel is from the same brain as the one below, but ~1,800 μm anterior to it. Scale bar represents 1 mm. d, Quantification of H&E staining, one-way ANOVA (n = 26 brain sections for all experimental groups; P = 2.9 × 10–14, F (2, 75) = 48.61) followed by Bonferroni’s post hoc test (WT versus KO, P = 4.9 × 10–14, 95% C.I. = 170.1–279.4, WT versus Het, P = 0.0678, 95% C.I. = –3.027–106.2). e, Coronal sections of E17.5 brains stained with antibodies against Mettl14 and Pax6. Similar results were obtained from three independent experiments. f, Coronal sections of E13.5, E15.5, E17.5 and P0 brains stained with anti-Mettl14. Similar results were obtained from three independent experiments. Scale bars represent 100 μm. Graphs represent the mean ± s.d. Dots represent data from individual data points. The horizontal lines in the box plots indicate medians, the box limits indicate first and third quantiles, and the vertical whisker lines indicate minimum and maximum values. ns, non-significant. ****P < 0.0001.
Next, we evaluated m6A function in isolated embryonic NSCs cultured in vitro. To determine which embryonic stages are appropriate to select Mettl14-deficient NSCs, we examined Mettl14 protein expression in coronal sections prepared from E13.5, E15.5, E17.5 and P0 brains from cKO, heterozygous and nondeleted control mice. Immunostaining revealed residual Mettl14 staining in the cerebral cortex at E13.5 in _Mettl14-_cKO brain, whereas Mettl14 signals in cortex were absent from E15.5 onward (Fig. 1f). Heterozygous mice showed Mettl14 signals comparable to those of nondeleted controls. Thus, for further analysis we chose E14.5 and E17.5 cortical NSCs and cultured them as neurospheres for 7 d before harvesting them for analysis.
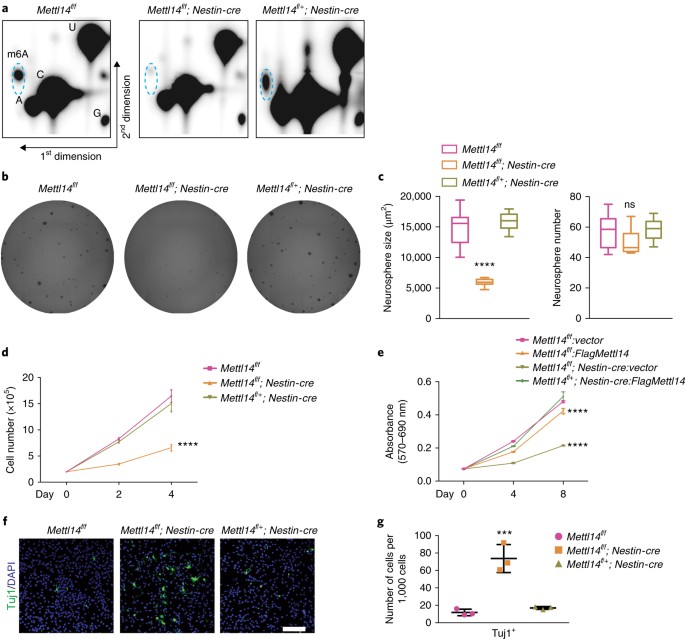
We observed comparable phenotypes in subsequent in vitro analysis of E14.5 and E17.5 NSCs. Unless stated otherwise, our results are those of experiments conducted in E14.5 NSCs. Following confirmation of Mettl14 loss in KO NSCs by western blotting (Supplementary Fig. 2a), we assessed m6A levels from E14.5 neurospheres. Thin-layer chromatography (TLC) analysis revealed an almost total loss of m6A in polyA RNA isolated from _Mettl14_-KO versus nondeleted NSCs, whereas heterozygous cells displayed m6A levels comparable to those seen in nondeleted controls (Fig. 2a), suggesting that the KO system that we generated is ideal for studying m6A function in NSCs.
Fig. 2: Mettl14 regulates self-renewal of cortical NSCs from E14.5 brain in neurosphere culture.
a, Two-dimensional thin-layer chromatography (2D-TLC) analysis of m6A levels in ribosome-depleted (Ribo-) PolyA RNAs isolated from E14.5 NSCs after 7 d of neurosphere culture. Dashed blue circles indicate m6A spots. Similar results were obtained from three independent experiments. b, Representative images of neurospheres formed from isolated E14.5 NSCs. c, Quantification of neurosphere number and area, one-way ANOVA (n = 12 cell cultures for all experimental groups; area, P = 9.15 × 10–13, F(2, 33) = 80.21; number, P = 0.0313, F(2, 33) = 3.853) followed by Bonferroni’s post hoc test (area, WT versus KO, P = 3.2475 × 10–11, 95% C.I. = 6,781–10,737, WT versus Het, P = 0.2855, 95% C.I. = –2,999–663.1; number, WT versus KO, P = 0.0724, 95% C.I. = –0.5596–15.39, WT versus Het, P = 0.9999, 95% C.I. = = –9.31–6.643). d, NSC growth curve. NSCs were plated at 200,000 cells per well in six-well plates and counted 2 and 4 d later; two-way ANOVA (n = 3 cell cultures for all experimental groups; P = 8.644 × 10–12, F(2, 18) = 143.6) followed by Bonferroni’s post hoc test (WT versus KO, P = 1.2905 × 10–11, 95% C.I. = 4.133–5.666, WT versus Het, P = 0.091, 95% C.I. = –0.09277–1.44). e, Growth curve of _Mettl14_-KO and nondeleted control NSCs transduced with indicated vectors. NSCs were plated in 96-well plates, and numbers were determined by MTT assay; two-way ANOVA (n = 3 cell cultures for all experimental groups; P = 1.413 × 10–20, F(3, 24) = 396.9) followed by Bonferroni’s post hoc test (WT-vector versus WT-FlagMettl14, P = 1.162 × 10–8, 95% C.I. = 0.02849–0.0514; WT-vector versus KO-vector, P = 1.7709 × 10–20, 95% C.I. = 0.1213–0.1442; WT-vector versus KO-FlagMettl14, P = 0.9999, 95% C.I. = –0.01183–0.01107). f, Immunostaining for anti-Tuj1 in NSCs cultured 7 d in vitro. Scale bar represents 100 μm. g, Quantification of immunostaining, one-way ANOVA (n = 3 fields for all experimental groups; P = 0.0004, F(2, 6) = 38.49) followed by Bonferroni’s post hoc test (WT versus KO, P = 0.0004, 95% C.I. = –85.13 to –38.65; WT versus Het, P = 0.9999, 95% C.I. = –28.37–18.11). Graphs represent the mean ± s.d. Dots represent data from individual data points. The horizontal lines in the box plots indicate medians, the box limits indicate first and third quantiles, and the vertical whisker lines indicate minimum and maximum values. ns, non-significant. ***P < 0.001, ****P < 0.0001.
To characterize KO versus control NSCs, we used a Celigo image cytometer and software to image neurospheres and assess their number and size. Although _Mettl14_-KO, heterozygous and nondeleted control NSCs derived from E14.5 embryos formed a similar number of neurospheres, neurosphere size, as reflected by neurosphere area in this system, decreased by ~55% in KO versus nondeleted control cells, whereas neurosphere size from heterozygous cells was comparable to that seen in nondeleted controls (Fig. 2b,c). Consistently, those same _Mettl14_-KO NSCs exhibited significantly decreased proliferation, as determined by cell-counting analysis (Fig. 2d). Similar proliferation defects were detected in NSCs taken from E17.5 _Mettl14_-cKO mice (Supplementary Fig. 2b). Annexin V flow cytometry (Supplementary Fig. 2c,d) and TUNEL (terminal deoxynucleotidyl transferase (TdT) dUTP nick-end labeling) analysis (Supplementary Fig. 2e) of E14.5 NSCs confirmed that the effects were not a result of increased apoptosis. To ensure that proliferation defects were a result of Mettl14 deletion, we performed rescue experiments by overexpressing Flag-tagged Mettl14 in E14.5 KO NSCs. Western and dot blot analysis confirmed Mettl14 transgene expression (Supplementary Fig. 2f) and the restoration of m6A (Supplementary Fig. 2g). Notably, Mettl14 overexpression did not increase the proliferation of nondeleted NSC controls (Fig. 2e), but increased the proliferation of _Mettl14_-KO NSCs to rates comparable to that of controls (Fig. 2e). These results suggest that Mettl14 and concomitant m6A RNA modification regulate NSC proliferation, at least in vitro.
It is well established that decreased NSC proliferation is coupled with premature NSC differentiation14. Thus, we checked for the presence of cells expressing the neuronal marker Tuj1 in E14.5 NSCs cultured for 7 d as neurospheres. Immunostaining analysis revealed a 6.2-fold increase in the number of Tuj1+ cells in KO versus control NSCs (Fig. 2f,g), whereas the number of Tuj1+ cells was comparable between heterozygous and nondeleted controls, suggesting that Mettl14 loss leads to premature neuronal differentiation. Together, these results suggest that Mettl14 regulates NSC self-renewal.
To determine whether the loss of an m6A demethylase would have the opposite effect on NSC proliferation, we knocked down two reported m6A demethylases, Fto and Alkbh59,15, separately in wild-type NSCs. Reverse-transcription quantitative PCR (RT-qPCR) analysis showed high knockdown efficiency in each case, and western blots revealed a marked decrease in Fto and Alkbh5 protein levels in the respective knockdown cells (Supplementary Fig. 2h,i,k,l). Nonetheless, NSC proliferation in vitro was not altered by the loss of either protein (Supplementary Fig. 2j,m). Some reports suggest that changes in expression of either Fto or Alkbh5 have only moderate effects on m6A levels and that both factors likely regulate m6A modification of a subset of transcripts15,16,17. Our results suggest that mRNAs that function in NSC proliferation are not regulated by either Fto or Alkbh5.
The size of the cortical RGC pool is reduced in _Mettl14_-cKO mouse brain
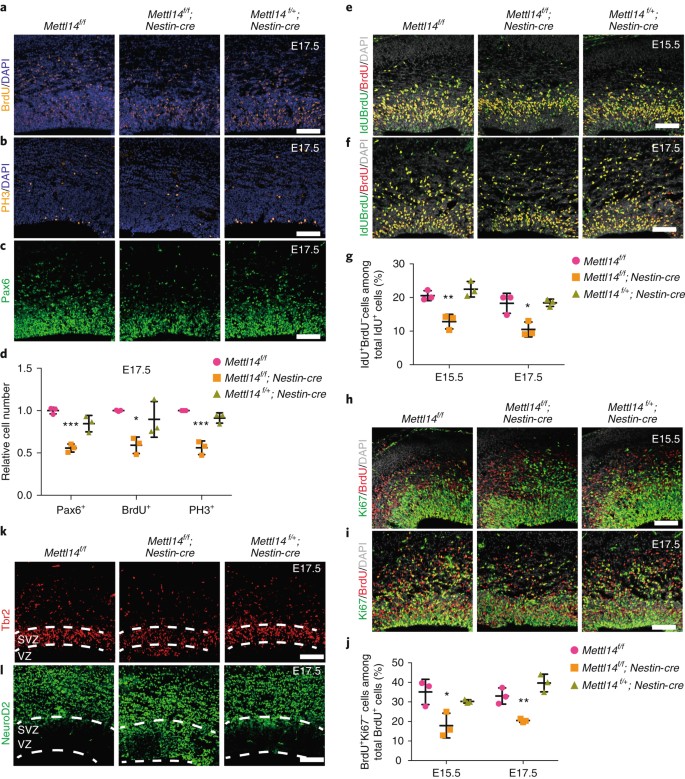
We next examined the effect of Mettl14 on the proliferation of primary cortical stem cells or RGCs in vivo. To do so, we determined the number of S-phase cells in the cortex of E13.5, E15.5 or E17.5 _Mettl14_-cKO, heterozygous and nondeleted control embryos by injecting pregnant females with bromodeoxyuridine (BrdU) and harvesting embryos 0.5 h later, which detected only cells undergoing DNA replication at that time point. Immunostaining showed that the number of BrdU+ cells decreased by 19% in E15.5 _Mettl14_-cKO compared with nondeleted control brain (Supplementary Fig. 3a,d), and that number was 40% when analysis was conducted at E17.5 (Fig. 3a,d). Similarly, we also observed a 44% and 45% decrease in the number of cells expressing the mitotic marker phospho-histone H3 (PH3) at the apical membrane of the cortical VZ in cKO versus nondeleted control brain at E15.5 and E17.5, respectively (Supplementary Fig. 3b,d and Fig. 3b,d). To determine the number of RGCs, we assessed brain coronal sections at all three stages with the RGC marker Pax6. Consistently, we detected a 45% decrease in the number of Pax6+ RGCs in the VZ of E17.5 _Mettl14_-cKO brain versus controls (Fig. 3c,d) and a 14% decrease at E15.5 (Supplementary Fig. 3c,d). All experiments showed highly comparable results between heterozygous and nondeleted control RGCs (Fig. 3a–d and Supplementary Fig. 3a–d). We did not detect differences in BrdU, PH3 or Pax6 staining relative to that in nondeleted controls in the cortex of E13.5 _Mettl14_-cKO brains (Supplementary Fig. 3e–g), consistent with the finding that residual Mettl14 is present in cortex at E13.5 (Fig. 1f). Immunostaining with the apoptosis marker cleaved caspase-3 revealed no change in the number of apoptotic cells in the cortex of E17.5 and E15.5 _Mettl14_-cKO brains relative to nondeleted controls (Supplementary Fig. 3h,i). To understand how Mettl14 loss might affect RGC proliferation, we examined cell cycle progression and cell cycle exit of RGCs from the brains of E15.5 and E17.5 _Mettl14_-cKO versus control mice. We carried out sequential 5-iodo-2ʹ-deoxyuridine (IdU) and BrdU injection to evaluate cell cycle progression, followed by IdU and BrdU double-staining of cortical sections18. We then determined the percentage of IdU+BrdU– cells, which represented the cells that had progressed past S-phase, versus all IdU+ cells, a group that included both proliferating cells and cells that had left S-phase. We detected a 38% and 43% decrease in E15.5 and E17.5 _Mettl14_-cKO embryos, respectively, compared with the nondeleted control, suggesting that Mettl14 loss disrupts normal RGC cell cycle progression (Fig. 3e–g). Heterozygous and nondeleted control RGCs yielded comparable results (Fig. 3e–g). To determine whether Mettl14 loss alters cell cycle exit, we performed BrdU-Ki67 double-staining of cortical sections from the brains of mice pulsed with BrdU and analyzed 24 h later. Mettl14 loss resulted in a 50% and 39% decrease in cells exiting the cell cycle from E15.5 and E17.5 _Mettl14_-cKO embryos, respectively, versus nondeleted controls, suggesting that Mettl14 is required for normal RGC cell cycle exit (Fig. 3h–j). Heterozygous and nondeleted control RGCs yielded comparable results (Fig. 3h–j). Together, these data strongly suggest that Mettl14 regulates the RGC cell cycle and that the RGC pool in cortex is substantially reduced in _Mettl14_-cKO mice.
Fig. 3: Mettl14 knockout decreases RGC proliferation in vivo.
a–c, Coronal sections of E17.5 brains stained with antibodies recognizing BrdU, PH3 and Pax6. Pregnant mothers received a BrdU pulse 30 min before embryo collection. d, Quantification of immunostaining from E17.5 sections. Numbers of Pax6+, BrdU+ and PH3+ cells were determined and normalized to those in comparable sections from nondeleted mice; one-way ANOVA (n = 3 brain sections for all experimental groups; Pax6+, P = 0.0005, F(2, 6) = 34.41; BrdU+, P = 0.0231, F(2, 6) = 7.531; PH3+, P = 0.0002, F(2, 6) = 47.73) followed by Bonferroni’s post hoc test (Pax6+, WT versus KO, P = 0.0004, 95% C.I. = 0.2814–0.6025, WT versus Het, P = 0.0584, 95% C.I. = –0.006443–0.3146; BrdU+, WT versus KO, P = 0.0194, 95% C.I. = 0.08378–0.7348, WT versus Het, P = 0.7612, 95% C.I. = –0.2218–0.4292; PH3+, WT versus KO, P = 0.0002, 95% C.I. = 0.2976–0.5796, WT versus Het, P = 0.2287, 95% C.I. = –0.05332–0.2288). e,f, Coronal sections of E15.5 (e) and E17.5 (f) brains stained with both anti-BrdU that recognizes BrdU only and anti-IdU that also recognizes BrdU. Pregnant mothers received one IdU injection, followed by one BrdU injection 1.5 h later. After another 0.5 h, the embryonic brains were collected for analysis. g, Quantification of the percentage of IdU+BrdU– cells, representing cells that left S-phase during the 1.5-h chase, among total IdU+ cells. One-way ANOVA (n = 3 brain sections for all experimental groups; E15.5, P = 0.0025, F(2, 6) = 19.21; E17.5, P = 0.0075, F(2, 6) = 12.35) followed by Bonferroni’s post hoc test (E15.5, WT versus KO, P = 0.0067, 95% C.I. = 2.835–12.6, WT versus Het, P = 0.5802, 95% C.I. = –6.787–2.973; E17.5, WT versus KO, P = 0.0107, 95% C.I. = 2.347–13.19, WT versus Het, P = 0.9999, 95% C.I. = –5.598–5.244). h,i, Coronal sections of E15.5 (h) and E17.5 (i) brains stained with antibodies recognizing Ki67 and BrdU. Pregnant mothers received one BrdU injection 24 h before embryo collection. j, Quantification of the percentage of BrdU+Ki67– cells, representing cells that exited the cell cycle during 24 h, among total BrdU+ cells. One-way ANOVA (n = 3 brain sections for all experimental groups; E15.5, P = 0.0173, F(2, 6) = 8.589; E17.5, P = 0.0016, F(2, 6) = 22.51) followed by Bonferroni’s post hoc test (E15.5, WT versus KO, P = 0.014, 95% C.I. = 4.493–29.92, WT versus Het, P = 0.6051, 95% C.I. = –7.885–17.54; E17.5, WT versus KO, P = 0.01, 95% C.I. = 3.932–21.2, WT versus Het, P = 0.1249, 95% C.I. = –15.28–1.99). k,l, Immunostaining of coronal sections of E17.5 brain with antibodies to the intermediate progenitor marker Tbr2 and the proneural marker Neurod2. Dashed white lines indicate border of VZ/SVZ area. Similar results were obtained from three independent experiments. Scale bars represent 100 μm. Graphs represent the mean ± s.d. Dots represent data from individual data points. ns, non-significant. *P < 0.05, **P < 0.01, ***P < 0.001.
We then examined RGC premature differentiation in the cortical VZ of E15.5 and E17.5 _Mettl14_-cKO brains using Eomes (Tbr2), a marker of intermediate progenitor cells located at the sub-ventricular zone (SVZ), and the proneural marker Neurod2 (ND2). Notably, patches of ND2+Tbr2+ cells were seen consistently at E15.5 and E17.5 in areas close to the apical surface of the cortical VZ of _Mettl14_-cKO brain, but were absent in comparably staged littermate controls (Supplementary Fig. 3j,k and Fig. 3k,l). Together, these data suggest that Mettl14 is required to prevent NSC premature differentiation and maintain the NSC pool in vivo.
Mettl14 deletion results in reduced numbers of late-born neurons
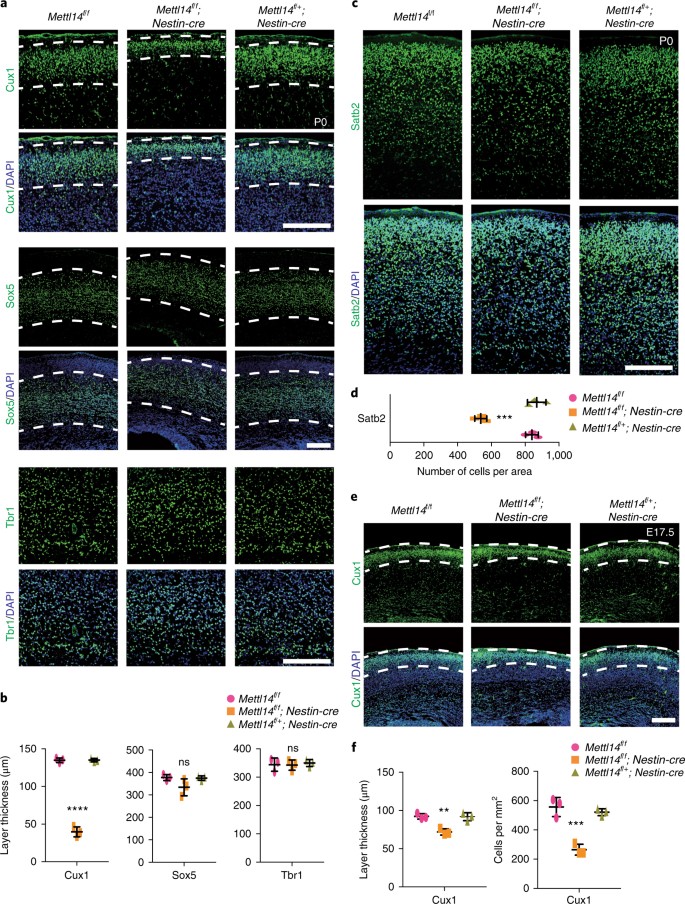
We next evaluated the effects of Mettl14 loss on cortical neurogenesis. In P0 mice, neurons differentiated from RGCs are found in six distinct cortical layers containing neuronal subtypes identifiable by specific markers. Thus, we stained coronal sections from cKO and comparably staged littermate controls at P0 for the following markers: Cux1, which is expressed in late-born neurons and is a marker of upper neuronal layers II–IV; Sox5, which is expressed in early-born neurons and is a marker of layer V; and Tbr1, which is expressed in postmitotic neurons and is a marker of layer VI to the subplate. Overall, layer organization was comparable in cKO and control mice. When we assessed layer thickness, the thickness of layers marked by Sox5 and Tbr1 did not differ significantly between genotypes (Fig. 4a,b). However, we observed a 70% decrease in the thickness of Cux1+ layers (II–IV) (Fig. 4a,b). To confirm the loss of neurons from these layers, we stained sections from P0 embryos for a different layer II–IV marker, Satb2, and observed an ~34% decrease in the number of Satb2+e neurons in cKO mice versus littermate controls (Fig. 4c,d). When we examined cortical Cux1 staining at E17.5, we detected a 22% reduction in the thickness of Cux1+ layers and a 50% reduction in the number of newly generated Cux1+ cells residing in a region between the VZ and layer IV in _Mettl14_-cKO mice versus controls (Fig. 4e,f). These results suggest that Mettl14 loss may deplete the progenitor pool in a way that is reflected by loss of late-born neurons.
Fig. 4: Mettl14 knockout decreases the number of late-born neurons.
a, Coronal sections of P0 brains stained for the layer II–IV marker Cux1, the layer V marker Sox5 and the layer VI/subplate (SP) marker Tbr1. Dashed white lines mark borders of Cux1+ and Sox5+ neuronal layers. b, Quantification of thickness of Cux1+, Sox5+ and Tbr1+ neuronal layers; one-way ANOVA (n = 3 brain sections for all experimental groups; Cux1+, P = 2.689 × 10–7, F(2, 6) = 461.8; Sox5+, P = 0.115, F(2, 6) = 3.169; Tbr1+, P = 0.8865, F(2, 6) = 0.1229) followed by Bonferroni’s post hoc test (Cux1+, WT versus KO, P = 4 × 10–7, 95% C.I. = 84.39–105.9, WT versus Het, P = 0.9999, 95% C.I. = –10.97–10.52; Sox5+, WT versus KO, P = 0.1329, 95% C.I. = –14.18–101.2, WT versus Het, P = 0.9999, 95% C.I. = –55.32–60.06; Tbr1+, WT versus KO, P = 0.9999, 95% C.I. = –43.31–46.59, WT versus Het, P = 0.9999, 95% C.I. = –50.47–39.42). c, Coronal sections of P0 brains stained for the layer II–IV marker Satb2. d, Quantification of the number of Satb2+ cells; one-way ANOVA (n = 3 brain sections for all experimental groups; P = 0.00015, F(2, 6) = 53.83) followed by Bonferroni’s post hoc test (WT versus KO, P = 0.0003, 95% C.I. = 198.2–408.5; WT versus Het, P = 0.9186, 95% C.I. = –133.1–77.14). e, Coronal sections of E17.5 brains stained for Cux1; dashed white lines mark the border of the Cux1+ neuronal layer. f, Quantification of Cux1+ layer thickness within dashed white lines and of the average number of newly generated Cux1+ cells within 1 mm2, as measured from the VZ to the lower dashed white lines, at E17.5. One-way ANOVA (n = 3 brain sections for all experimental groups; thickness, P = 0.0019, F(2, 6) = 21.36; number, P = 0.0004, F(2, 6) = 36.75) followed by Bonferroni’s post hoc test (thickness, WT versus KO, P = 0.0025, 95% C.I. = 9.765–30.85, WT versus Het, P = 0.9999, 95% C.I. = –10.13–10.96; number, WT versus KO, P = 0.0004, 95% C.I. = 181.5–401.9, WT versus Het, P = 0.7499, 95% C.I. = –74.64–145.8). Scale bars represent 200 μm. Graphs represent the mean ± s.d. Dots represent data from individual data points. ns, non-significant. **P < 0.01, ***P < 0.001, ****P < 0.0001.
Mettl14 knockout leads to genome-wide changes in histone modification that perturb gene expression
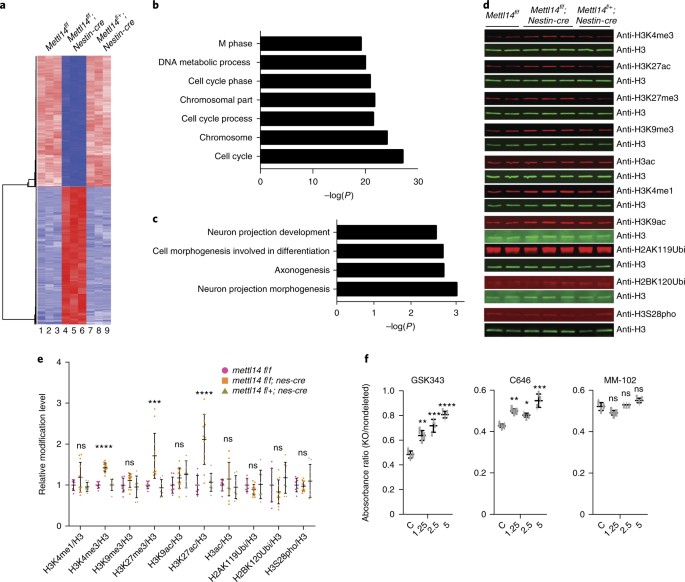
To assess molecular mechanisms underlying m6A-regulated NSC activity, we cultured NSCs from E14.5 _Mettl14_-cKO, heterozygous and nondeleted control embryos for 7 d and performed RNA sequencing (RNA-seq). _Mettl14_-KO NSCs exhibited distinct gene expression profiles relative to nondeleted and heterozygous controls (which showed comparable profiles; Fig. 5a and Supplementary Table 3). Gene Ontology analysis (GO) suggested that the most significantly upregulated genes function in NSC differentiation, whereas downregulated genes are associated with cell proliferation (Fig. 5b,c and Supplementary Fig. 4a), changes that are reflective of observed phenotypes. We then evaluated potential mechanisms underlying these changes in gene expression. It is well established that m6A destabilizes transcripts4,13,19,20. However, we detected only a weak correlation between m6A loss and increase in transcript abundance (Supplementary Fig. 4b, Supplementary Tables 3 and 4), suggesting that different m6A-related mechanisms modulate mRNA levels. Given that modification of histone tails is a critical mechanism of gene regulation in mammalian cells21, we asked whether m6A RNA modification may also change histone modifications by performing western blotting on acid-extracted histones from KO versus control NSCs isolated at both E14.5 and E17.5. We evaluated a panel of well-studied histone modifications that have been reported to regulate stem cell activities, including histone H3 phosphorylation, histone H2A and H2B ubiquitination, three types of histone acetylation, and four types of histone methylation22,23,24,25,26,27,28,29,30,31,32,33. These histone marks are associated with either gene activation or repression. Representative western blots of E14.5 NSCs are shown in Fig. 5d. We quantified western blots from E14.5 and E17.5 by calculating the ratios of respective histone modifications to total H3 protein in KO, heterozygous and nondeleted control NSCs. Although we observed no significant change in any of the histone modifications that we tested between heterozygous and nondeleted control samples (Fig. 5d,e), we detected a significant increase in acetylation of histone H3 at lysine 27 (H3K27ac; 111% increase), trimethylation of histone H3 at lysine 4 (H3K4me3; 43% increase) and trimethylation of histone H3 at lysine 27 (H3K27me3; 71% increase) in _Mettl14_-KO NSCs versus nondeleted controls (Fig. 5d,e). These results suggest that m6A regulates specific histone modifications.
Fig. 5: m6A regulates NSC gene expression through histone modifications.
a, Heat map analysis based on RNA-seq analysis in _Mettl14_-KO versus nondeleted control NSCs. b,c, GO analysis of genes down- and upregulated in _Mettl14_-KO versus nondeleted control E14.5 NSCs. GO analyses were performed by DAVID. Differentially expressed genes had an adjusted P < 0.01 and a twofold or greater expression difference. Among differentially expressed genes, 1,099 are upregulated and 1,487 are downregulated. Numbers of gene counts and exact P values for each GO term are listed in Supplementary Fig. 4a. d, Representative western blots of acid-extracted histones from E14.5 NSCs using antibodies recognizing H3K4me1, H3K4me3, H3K27me3, H3K9me3, H3K27ac, H3K9ac, H3ac, H2AK119Ubi, H2BK120Ubi and H3S28pho. The band sizes range from 17 to 23 kDa as expected for modified histones. For uncropped images, see Supplementary Fig. 6a. e, Quantitation of western blots from E14.5 and E17.5 NSCs. One-way ANOVA (n = 8 (WT), 12 (KO), or 8 (Het) independent NSC cultures; H3K4me1, P = 0.1123, F(2, 25) = 2.39; H3K4me3, P = 1.06442 × 10–9, F(2, 25) = 52.77; H3K9me3, P = 0.2096, F(2, 25) = 1.664; H3K27me3, P = 0.00013, F (2, 25) = 13.07; H3K9ac, P = 0.1461, F(2, 25) = 2.08; H3K27ac, P = 4.796 × 10–6, F(2, 25) = 20.8; H3ac, P = 0.3676, F(2, 25) = 1.042; H2AK119Ubi, P = 0.3592, F(2, 25) = 1.067; H2BK120Ubi, P = 0.1192, F(2, 25) = 2.319; H3S28pho, P = 0.5347, F(2, 25) = 0.642) followed by Bonferroni’s post hoc test (H3K4me1, WT versus KO, P = 0.2376, 95% C.I. = –0.4713–0.09065, WT versus Het, P = 0.9999, 95% C.I. = –0.2629–0.3527; H3K4me3, WT versus KO, P = 1.157 × 10–8, 95% C.I. = –0.5518 to –0.3128, WT versus Het, P = 0.9999, 95% C.I. = –0.134–0.1278; H3K9me3, WT versus KO, P = 0.4574, 95% C.I. = –0.3314–0.1054, WT versus Het, P = 0.9999, 95% C.I. = –0.1942–0.2842; H3K27me3, WT versus KO, P = 0.0008, 95% C.I. = –1.131 to –0.2956, WT versus Het, P = 0.9999, 95% C.I. = –0.3891–0.5256; H3K9ac, WT versus KO, P = 0.321, 95% C.I. = –0.4577–0.1121, WT versus Het, P = 0.1141, 95% C.I. = –0.5732–0.05098; H3K27ac, WT versus KO, P = 1.769 × 10–5, 95% C.I. = –1.591 to –0.6358, WT versus Het, P = 0.9999, 95% C.I. = –0.5908–0.4556; H3ac, WT versus KO, P = 0.6463, 95% C.I. = –0.4945–0.2007, WT versus Het, P = 0.9999, 95% C.I. = –0.3309–0.4307; H2AK119Ubi, WT versus KO, P = 0. 5288, 95% C.I. = –0.1242–0.3523, WT versus Het, P = 0.9999, 95% C.I. = –0.2759–0.2459; H2BK120Ubi, WT versus KO, P = 0.6171, 95% C.I. = –0.2165–0.5511, WT versus Het, P = 0.6457, 95% C.I. = –0.5982–0.2426; H3S28pho, WT versus KO, P = 0.9999, 95% C.I. = –0.2407–0.2961, WT versus Het, P = 0.8731, 95% C.I. = –0.3915–0.1965). f, Cell growth analysis based on an MTT assay of NSCs treated with vehicle/DMSO or the MLL1 inhibitor MM-102, the CBP/P300 inhibitor C646, or the Ezh2 inhibitor GSK343. Shown is the absorbance ratio of KO to nondeleted controls at each drug dose. One-way ANOVA (n = 3 independent experiments for all experimental groups; GSK343, P = 4.232 × 10–5, F(3, 8) = 38.47; C646, P = 0.0003, F(3, 8) = 23.43; MM-102, P = 0.0025, F(3, 8) = 11.91) followed by Bonferroni’s post hoc test (GSK343, c versus 1.25, P = 0.0035, 95% C.I. = –0.2477 to –0.05943, c versus 2.5, P = 0.0002, 95% C.I. = –0.3265 to –0.1383, c versus 5, P = 1.979 × 10–5, 95% C.I. = –0.4169 to –0.2287; C646, c versus 1.25, P = 0.0036, 95% C.I. = –0.1158 to –0.02744, c versus 2.5, P = 0.0236, 95% C.I. = –0.09574 to –0.007344, c versus 5, P = 0.000103, 95% C.I. = –0.1654 to –0.07702; MM-102, c versus 0.0625, P = 0.0507, 95% C.I. = –8.591 × 10–5 to 0.06086, c versus 1.25, P = 0.9999, 95% C.I. = –0.03858–0.02237, c versus 2.5, P = 0.0615, 95% C.I. = –0.05958–0.001368). Graphs represent the mean ± s.d. Dots represent data from individual data points. ns, non-significant. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To determine whether these changes alter NSC proliferation, we searched for chemical inhibitors that antagonize activities associated with upregulated histone modifications to determine whether inhibitor treatment of E14.5 KO NSCs would rescue cell proliferation defects. Three inhibitors were commercially available: MM102, which inhibits mixed-lineage leukemia (MLL) function and H3K4me3 formation; C646, which inhibits the H3K27 acetyltransferase Crebbp (CBP)/p300 activity; and GSK343, which inhibits Ezh2-dependent H3K27me3 formation. We then seeded comparable numbers of NSCs of all three genotypes, added inhibitor or DMSO vehicle at day 0, and determined cell number via MTT assays 4 d later. After DMSO treatment, the number of KO NSCs was ~50% that of nondeleted controls, reflecting slower proliferation, as anticipated (Fig. 5f). GSK343 treatment at 1.25, 2.5 and 5 μM increased the ratios of KO to nondeleted control NSCs to 64%, 71% and 80%, respectively (Fig. 5f), whereas the percentages of heterozygous to nondeleted control NSCs were unchanged by GSK343 treatment (Supplementary Fig. 4c). These observations suggest that blocking the formation of H3K27me3 rescues the growth defects of KO NSCs. Increased ratios of KO versus nondeleted control NSCs were also seen after C646 treatment (Fig. 5f and Supplementary Fig. 4c), suggesting that blocking of H3K27ac also blocks the proliferation defects of KO NSCs. By contrast, treatment of E14.5 NSCs with MM102 had no effect (Fig. 5f and Supplementary Fig. 4c). These results suggest that m6A regulates NSC proliferation, at least in part, through H3K27me3 and H3K27ac modifications.
H3K27me3 marks gene promoters and is associated with silencing34,35, whereas H3K27ac, which is enriched at promoters and enhancers, is associated with gene activation36,37. Thus, we asked whether increased promoter H3K27me3 was associated with gene downregulation, whereas increased promoter/enhancer H3K27ac was associated with gene upregulation in E14.5 _Mettl14_-KO versus control NSCs. To do so, we performed H3K27me3 and H3K27ac ChIP-seq analysis on E14.5 KO versus nondeleted NSCs (Supplementary Tables 5 and 6) and correlated changes in gene expression with altered histone modification. In total, the intensity of 1,610 promoter/enhancer H3K27ac peaks, defined as peaks within a 10-kb region up- or downstream of a transcriptional start site (TSS), were significantly altered in KO versus control cells. Pearson correlation analysis showed a positive correlation between changes in peak intensity and changes in gene expression (r = 0.06195, P = 0.01292) in KO versus control NSCs, suggesting that H3K27ac functions in m6A-regulated gene activation. We also detected 434 altered H3K27me3 promoter peaks, defined as peaks within 2 kb upstream of a TSS, in KO versus control NSCs. Although in this case we did not detect a significant correlation between changes in peak intensity and gene expression (P = 0.05784) using all 434 genes, we detected a strongly negative Pearson correlation (r = –0.38804, P < 0.02) when we analyzed only downregulated genes (log2 fold change ≤ –0.6) in KO versus control NSCs, suggesting that H3K27me3 levels are positively correlated to the repression of genes showing decreased expression.
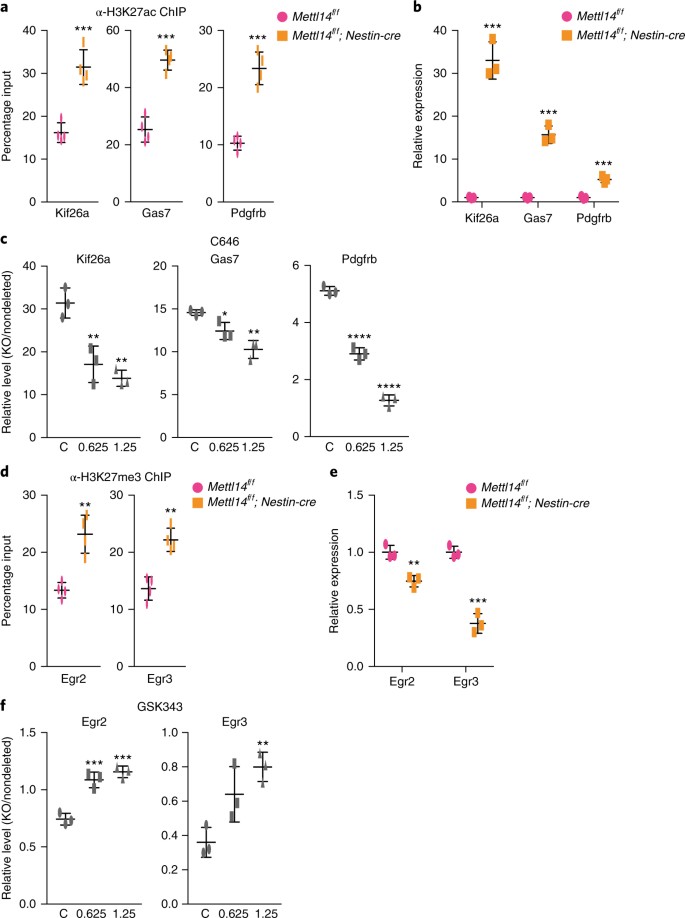
To further assess the relevance of altered H2K27ac and H3K27me3 modifications to NSC gene expression, we asked whether altered transcript abundance seen in KO versus control NSCs could be rescued by treating cells with inhibitors of H2K27ac (C646) or of H3K27me3 (GSK343). Using overlaying ChIP-seq and RNA-seq data and coupling that to Ingenuity pathway analysis (IPA), we picked five differentiation-related genes showing increased H3K27ac and increased expression and five cell-proliferation related genes showing increased H3K27me3 but decreased expression for rescue experiment. Indeed, C646 treatment resulted in significantly decreased expression of the neuritogenesis regulators Kif26a38, Gas739 and Pdgfrb40, in KO NSCs when compared with that in nondeleted NSCs (Fig. 6a–c), whereas GSK343 treatment increased expression of the transcription factors Egr2 and Egr3, which are known to promote proliferation41 (Fig. 6d–f). These results suggest that m6A-regulated histone modification functions in NSC gene expression.
Fig. 6: H3K27ac inhibitor C646 and H3K27me3 inhibitor GSK343 rescue aberrant gene expression in KO versus nondeleted NSCs.
a, H3K27ac ChIP-qPCR showing increased promoter and enhancer H3K27ac of Kif26a, Gas7 and Pdgfrb genes in E14.5 _Mettl14_-KO versus nondeleted NSCs. n = 4 independent experiments for all experimental groups; two-tailed unpaired _t_-test (Kif26a, P = 0.0006, t = 6.568, df = 6, 95% C.I. = 9.594–20.99; Gas7, P = 0.00013, t = 8.638, df = 6, 95% C.I. = 17.41–31.16; Pdgfrb, P = 0.0002, t = 8.395, df = 6, 95% C.I. = 9.27–16.9). b, RT-qPCR showing increased expression of Kif26a, Gas7 and Pdgfrb genes in E14.5 _Mettl14_-KO versus nondeleted NSCs. n = 3 independent experiments for all experimental groups; two-tailed unpaired _t_-test (Kif26a, P = 0.0002, t = 12.71, df = 4, 95% C.I. = 25.01–38.99; Gas7, P = 0.0002, t = 12.46, df = 4, 95% C.I. = 11.41–17.95; Pdgfrb, P = 0.0008, t = 9.08, df = 4, 95% C.I. = 2.957–5.563). c, RT-qPCR showing decreased expression of Kif26a, Gas7 and Pdgfrb genes in E14.5 _Mettl14_-KO versus nondeleted NSCs treated with H3K27ac inhibitor C646. One-way ANOVA (n = 3 independent experiments for all experimental groups; Kif26a, P = 0.0015, F(2, 6) = 23.04; Gas7, P = 0.0027, F(2, 6) = 18.67; Pdgfrb, P = 8.449 × 10–7, F(2, 6) = 314.3) followed by Bonferroni’s post hoc test (Kif26a, c versus 0.625, P = 0.0041, 95% C.I. = 6.126–22.47, c versus 1.25, P = 0.0014, 95% C.I. = 9.393–25.73; Gas7, c versus 0.625, P = 0.045, 95% C.I. = 0.05735–4.229, c versus 1.25, P = 0.0018, 95% C.I. = 2.207–6.379; Pdgfrb, c versus 0.625, P = 1.431 × 10–5, 95% C.I. = 1.75–2.663, c versus 1.25, P = 5.418 × 10–7, 95% C.I. = 3.384–4.296). d, H3K27me3 ChIP-qPCR showing increased H3K27me3 at promoters of Egr2 and Egr3 genes in E14.5 _Mettl14_-KO versus nondeleted NSCs. n = 4 independent experiments for all experimental groups; two-tailed unpaired _t_-test (Egr2, P = 0.0016, t = 5.463, df = 6, 95% C.I. = 5.412–14.19; Egr3, P = 0.0010, t = 5.928, df = 6, 95% C.I. = 5.007–12.05). e, RT-qPCR showing decreased expression of Egr2 and Egr3 genes in E14.5 _Mettl14_-KO versus nondeleted NSCs. n = 3 independent experiments for all experimental groups; two-tailed unpaired _t_-test (Egr2, P = 0.0052, t = 5.603, df = 4, 95% C.I. = –0.3789 to –0.1278; Egr3, P = 0.0009, t = 10.67, df = 4, 95% C.I. = –0.7855 to –0.4612). f, RT-qPCR showing increased expression of Egr2 and Egr3 genes in E14.5 _Mettl14_-KO versus nondeleted NSCs treated with H3K27me3 inhibitor GSK343. One-way ANOVA (n = 3 independent experiments for all experimental groups; Egr2, P = 0.0003, F(2, 6) = 44.49; Egr3, P = 0.01, F(2, 6) = 10.94) followed by Bonferroni’s post hoc test (Egr2, c versus 0.625, P = 0.0007, 95% C.I. = –0.4826 to –0.2041, c versus 1.25, P = 0.0002, 95% C.I. = –0.5526 to –0.2741; Egr3, c versus 0.625, P = 0.0519, 95% C.I. = –0.5627–0.002676, c versus 1.25, P = 0.0072, 95% C.I. = –0.7227 to –0.1573). Graphs represent the mean ± s.d. Dots represent data from individual data points. ns, non-significant. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
m6A regulates the stability of CBP and p300 transcripts
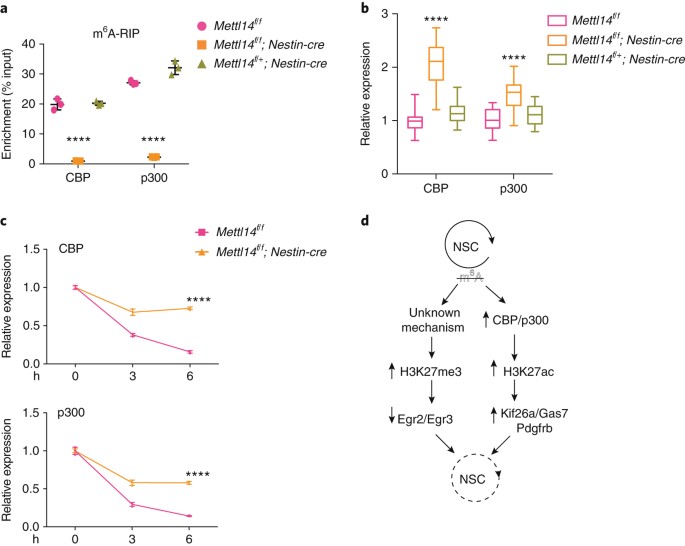
We then asked how m6A might regulate histone modifications. To do so, we first evaluated the presence of m6A on transcripts encoding the H3K27 acetyltransferases CBP and p300 and the polycomb repressive complex (PRC2) subunits Ezh2, Suz12 and Eed, which catalyze H3K27me3, by methylated RNA immunoprecipitation (meRIP). We detected a 20–30% enrichment of m6A over input in CBP (Crebbp) and p 300 (Ep300) mRNAs, which was lost in E14.5 _Mettl14_-KO NSCs (Fig. 7a). In contrast, only a 0.4–0.6% enrichment of m6A was observed in Ezh2, Eed and Suz12 mRNAs, and the extremely low levels that we observed for Ezh2 and Eed persisted in KO cells (Supplementary Fig. 5), suggesting that the signals that we detected are a result of the immunoprecipitation background.
Fig. 7: m6A regulates mRNA stability of CBP and p300.
a, m6A-meRIP-qPCR of CBP and p300 in _Mettl14_-KO versus control E14.5 NSCs. One-way ANOVA (n = 3 independent experiments for all experimental groups; CBP, P = 1.08 × 10–6, F(2, 6) = 289.4; p300, P = 3.961 × 10–7, F(2, 6) = 405.5) followed by Bonferroni’s post hoc test (CBP, WT versus KO, P = 1.697 × 10–6, 95% C.I. = 16.18–21.63, WT versus Het, P = 0.9999, 95% C.I. = –3.13–2.316; p300, WT versus KO, P = 1.113 × 10–6, 95% C.I. = 21.47–28.12, WT versus Het, P = 0.0086, 95% C.I. = –8.32 to –1.667). b, RT-qPCR of CBP and p300 transcripts in E14.5 NSCs cultured for 7 d in vitro; one-way ANOVA (n = 21 (WT), 33 (KO), or 21 (Het) independent experiments for all experimental groups; CBP, P = 2.380 × 10–21, F(2, 72) = 98.64; p300, P = 2.751 × 10–9, F(2, 72) = 26.24) followed by Bonferroni’s post hoc test (CBP, WT versus KO, P = 1.306 × 10–19, 95% C.I. = –1.252 to –0.8628, WT versus Het, P = 0.3029, 95% C.I. = –0.3512–0.07886; p300, WT versus KO, P = 5.254 × 10–9, 95% C.I. = –0.6356 to –0.3153, WT versus Het, P = 0.2011, 95% C.I. = –0.3058–0.04839). c, RT-qPCR of CBP and p300 transcripts in ActD-treated E14.5 NSCs. P values are generated by two-way ANOVA (n = 3 independent experiments for all experimental groups; CBP, P = 1.262 × 10–11, F(1, 12) = 602.5; p300, P = 8.738 × 10–10, F(1, 12) = 291.7) followed by Bonferroni’s post hoc test (CBP, 0 h, P = 0.9999, 95% C.I. = –0.05658–0.05658, 3 h, P = 1.714 × 10–8, 95% C.I. = –0.3518 to –0.2386, 6 h, P = 7.954 × 10–12, 95% C.I. = –0.6268 to –0.5136; p300, 0 h, P = 0.9999, 95% C.I. = –0.06777–0.06777, 3 h, P = 1.988 × 10–7, 95% C.I. = –0.3522 to –0.2167, 6 h, P = 1.50564 × 10–9, 95% C.I. = –0.5046 to –0.3691). d, A model in which m6A loss alters histone modifications partly through regulation of mRNA stability of histone modifiers, and altered histone modifications aberrantly repress proliferation-related genes and activate differentiation-related genes, resulting in loss of NSC ground state. Graphs represent the mean ± s.d. Dots represent data from individual data points. The horizontal lines in the box plots indicate medians, the box limits indicate first and third quantiles, and the vertical whisker lines indicate minimum and maximum values. ****P < 0.0001.
We then evaluated potential changes in the stability of CBP and p300 mRNAs. We observed a significant increase in both CBP and p300 mRNA levels in E14.5 _Mettl14_-KO versus control NSCs (Fig. 7b). We then assayed mRNA stability by treating E14.5 cultured KO and control NSCs with actinomycin D (ActD) to block transcription and harvesting cells 3 and 6 h later. Both CBP and p300 showed significantly increased mRNA stability in _Mettl14_-KO NSCs compared with nondeleted control NSCs (Fig. 7c), suggesting that m6A may regulate histone modification by destabilizing transcripts that encode histone modifiers.
Discussion
By conditionally inactivating Mettl14 in embryonic NSCs, we discovered that Mettl14 is required for NSC proliferation and maintains NSCs in an undifferentiated state (Figs. 1–3). Thus, our findings reveal a previously unknown, but essential, function of m6A RNA methylation in the regulation of NSC self-renewal. We also observed decreased numbers of late-born neurons, which are generated from RGCs after E15.5, in the cortex of _Mettl14_-cKO animals at E17.5 and P0 (Fig. 4), consistent with the loss of Mettl14 expression and a decrease in size of the RGC pool. Notably, although Mettl14 loss promoted premature NSC differentiation, the identity of neuronal subtypes in each neuronal layer was not obviously affected in cortex of _Mettl14_-cKO brain. Thus, we conclude that RGCs lacking Mettl14 remain capable of differentiation and migration, and propose that cortical defects seen in _Mettl14_-KO mice are primarily a result of perturbed NSC self-renewal. Overall, our results provide a benchmark to further explore mechanisms underlying perturbed m6A RNA methylation in neurodevelopmental disorders. We also detected Mettl14 expression in postmitotic cortical neurons (Fig. 1e); thus, the possibility of a Mettl14 function in these cells cannot be excluded. However, given that the _Mettl14_-cKO mice in our study died shortly after birth, examination of potential Mettl14 function in neurons at later postnatal or adult stages was not possible. We also note that, although we found a specific reduction in the number of upper layer projection neurons in the cortex of _Mettl14_-cKO versus control mice, we do not exclude the possibility that Mettl14 regulates production of neurons in other cortical layers. Analysis of that effect is likely to require the use of a Cre driver to delete Mettl14 at an earlier time point than E13.5.
Our data suggest, for the first time, to our knowledge, the existence of cross-talk between RNA and histone modification. Although m6A reportedly regulates gene expression through diverse mechanisms such as mRNA stability4,13,19,20, splicing15,42,43,44 and translation45,46, an interaction between m6A modification and epigenetic mechanisms has not been explored. Thus, our finding that m6A RNA methylation regulates specific histone modifications, including H3K27me3, H3K27ac and H3K4me3, represents a previously unknown mechanism of gene regulation. Among these modifications, H3K27ac and H3K4me3 are associated with gene activation, and H3K27me3e with repression, consistent with our observations that there is no marked bias toward gene activation or repression in _Mettl14_-KO NSCs compared with controls (Fig. 5a). Importantly, our data show that, in _Mettl14_-KO cells, different m6A-regulated histone marks regulate expression of genes of different function (Fig. 6), suggesting that m6A-regulated active and repressive histone modifications work synergistically to ensure an NSC ground state. Synergy is also evident in our observation that treatment of cells with either H3K27me3 or H3K27ac inhibitors can, at least partially, restore cell proliferation in _Mettl14_-KO NSCs (Fig. 5). We did not observe any effects of the H3K4me3 inhibitor MM-102 on NSC proliferation. However, unlike H3K27ac and H3K27me3, which are regulated by distinct enzymes, H3K4me3e modification is catalyzed by a panel of SET domain proteins47. Although MM-102 is widely used to test H3K4me3 function, it inhibits the formation of a MLL1–WDR5 complex and therefore targets only the subset of H3K4me3 modifications that are dependent on MLL148. As yet, there are no inhibitors available that inhibit activities of most H3K4me3 methyltransferases and therefore sufficiently abolish H3K4me3. Thus, we cannot exclude the possibility that m6A-regulated H3K4me3 regulates NSC proliferation. Future studies may reveal that m6A regulates levels of histone modifications other than H3K27ac, H3K27me3 and H3K4me3.
We further showed that m6A regulates histone modifications directly by destabilizing transcripts that encode histone modifiers. It is evident that such a mechanism is applicable to the H3K27 acetyltransferase CBP/p300, but not to subunits in the PRC2 complex, suggesting that m6A regulates histone modifications through distinct mechanisms. Identification of those mechanisms warrants future investigation.
In summary, we propose a model (Fig. 7d) in which m6A loss alters histone modifications partly by regulating mRNA stability of histone modifiers, and altered histone modifications aberrantly repress proliferation-related genes and activate differentiation-related genes, resulting in a loss of NSC ground state. Our results provide an in-depth analysis of m6A in brain neural stem cells and suggest that there is an interaction between m6A and histone modification as a mechanism of gene regulation.
Methods
Mettl14 gene targeting
Mettl14 gene targeting vector was created by E. coli bacterial recombination as described49. NotI-ApaI fragment including intron 1 to 5 of Mettl14 in the fosmid WI1-205L1 (CHORI) was cloned into pGEM11-DTA2L vector50, yielding pGEM-Mettl14(1-5)-DTA. Two primer pairs (M14-L50F and M14-L50R, the left arm for bacterial recombination; LoxP-LF and LoxP-LR) were annealed and cloned into XhoI/HindIII of pBS-2xFRT-Zeo, derivative of pBS-2xF5-Zeo51. The PCR fragment including Mettl14 intron 1 to 2 with loxP site (primer pair, M14-ex2F and M14-ex2R-LoxP) digested by BstBI-MluI and right arm adaptor (annealed primer pair, Mettl14-R50F and Mettl14-R50R) were cloned into the vector with the left arm. The splice acceptor (SA)-internal ribosomal entry site (Ires)-hygromycin resistance gene (Hyg)-tandem polyadenylation signals (tpA) cassette of pGEM-SA-Ires-Hyg-tpA was cloned into NheI-PmlI of the vector with left and right arms51. The bacterial targeting vector was digested by AgeI- NotI to remove vector backbone and was transfected into E. coli strain SW106 containing pGEM-Mettl14(1-5)-DTA for bacterial recombination, yielding Mettl14 targeting vector for mouse ES cells.
Mettl14 ES targeting vector was linearized by NotI and transfected into G4 male ES cells as described52,53. 24 h after transfection, 150 μg/ml Hygromycin was added to the ES medium. Hygromycin-resistant ES colonies were picked 10 d after transfection. Targeting of Mettl14 was confirmed by genomic PCR analysis using primer pairs: M14cF and SA-R for 5′-end; UptpA-F and M14-cR for 3′-end.
Generation of Mettl14 conventional and conditional knockout mice
Positive ES clones were used for injection into c57 blastocysts and generation of chimerical mice. To produce Mettl14 f/+ mice, the chimeras were crossed with wild-type c57 for germ line transmission and then crossed with Atcb-Flpe transgenic mice (The Jackson Laboratory, # 003800) to remove FRT flanked selection cassette. Male Mettl14 f/+ mice were crossed to female EIIa-Cre transgenic mice (The Jackson Laboratory, # 003724) to obtain Mettl14 +/− mice, and Mettl14 +/− mice were intercrossed to obtain _Mettl14_–conventional knockout mice. Sex of embryos was determined.
To conditionally knock out Mettl14 in brain, floxed Mettl14 mice were bred with Nestin-Cre transgenic mice (The Jackson Laboratory, # 003771) to generate Mettl14 f/f ;Nestin-cre. Sex of embryos was not determined. Mice were maintained at the Sanford Burnham Prebys Medical Discovery Institute Institutional animal facility, and experiments were performed in accordance with experimental protocols approved by local Institutional Animal Care and Use Committees.
Genotyping
Genotyping was performed using the MyTaq Extract-PCR kit (Bioline) with primer sets corresponding to the primer list table (Supplementary Table 7).
Injection of S-phase tracer
IdU/BrdU double-labeling was performed as previously described18. Briefly, pregnant females were injected intraperitoneally with Iododeoxyuridine (IdU, Sigma) (100 mg/kg body weight) and 1.5 h later with the same dose of BrdU (Sigma) and killed after 0.5 h.
For BrdU single-labeling, pregnant females were injected intraperitoneally with BrdU (100 mg/kg body weight) and killed after 0.5 or 24 h.
Immunohistochemistry
Frozen sections were boiled in citrate buffer, pH 6.0 (Sigma), penetrated in 0.25% Triton X-100 in PBS for 30 min at room temperature and blocked in PBST (PBS with 0.2% Tween-20) containing 10% normal goat serum (Abcam). Sections were incubated with primary antibody at 4 °C overnight. Secondary antibodies were applied to sections for 2 h at room temperature. The primary antibodies used were as follows: rabbit anti-Mettl14 (1:500; Sigma, Cat. # HPA038002; immunohistochemistry validation and peer-reviewed citations at https://www.sigmaaldrich.com/catalog/product/sigma/hpa038002?lang=en®ion=US), Rabbit anti-Satb2 (1:500, Abcam, Cat. # Ab92446; immunohistochemistry validation and peer-reviewed citations at http://www.abcam.com/satb2-antibody-epncir130a-ab92446.html), Rabbit anti-Tbr1 (1:200, Abcam, Cat. # Ab31940; immunohistochemistry validation and peer-reviewed citations at http://www.abcam.com/tbr1-antibody-ab31940.html), rabbit anti-Sox5 (1:200; Abcam, Cat. # Ab94396; immunohistochemistry validation and peer-reviewed citations at http://www.abcam.com/sox5-antibody-chip-grade-ab94396.html), rabbit anti-Cux1 (1:100; Santa Cruz, Cat. # sc-13024; immunohistochemistry validation and peer-reviewed citations at https://www.scbt.com/scbt/product/cdp-antibody-m-222), rabbit anti-Pax6 (1:300; Biolegend, Cat. # PRB-278P; immunohistochemistry validation and peer-reviewed citations at https://www.biolegend.com/en-us/products/purified-anti-pax-6-antibody-11511), rat anti-BrdU (1:200; Abcam, Cat. # Ab6326; immunohistochemistry validation and peer-reviewed citations at http://www.abcam.com/brdu-antibody-bu175-icr1-ab6326.html), mouse anti-IdU/BrdU (1:50; BD bioscience, Cat. # 347580; immunohistochemistry validation and peer-reviewed citations at https://www.bioz.com/result/mouse%20anti%20brdu/product/BD%20Biosciences/?r=4.31&cf=0&uq=BrdU%20clone%20B44), rabbit anti-Ki67 (1:400, Cell Signaling, Cat. # 12202; immunohistochemistry validation and peer-reviewed citations at https://www.cellsignal.com/products/primary-antibodies/ki-67-d3b5-rabbit-mab-mouse-preferred-ihc-formulated/12202), rat anti-phospho histone 3 (1:300; Abcam, Cat. # Ab10543; immunohistochemistry validation and peer-reviewed citations at http://www.abcam.com/histone-h3-phospho-s28-antibody-hta28-ab10543.html), rabbit anti-Cleaved Caspase-3 (1:600; Cell Signaling, Cat. # 9661; immunohistochemistry validation and peer-reviewed citations at https://www.cellsignal.com/products/primary-antibodies/cleaved-caspase-3-asp175-antibody/9661), rabbit anti-NeuroD2 (1:1,000; Abcam, Cat. # Ab104430; immunohistochemistry validation and peer-reviewed citations at http://www.abcam.com/neurod2-antibody-ab104430.html), rabbit anti-Tbr2 (1:500, Abcam, Cat. # Ab23345; immunohistochemistry validation and peer-reviewed citations at http://www.abcam.com/tbr2-eomes-antibody-chip-grade-ab23345.html), mouse anti-Tuj1 (1:1,000; Stemcell Tech., Cat. # 01409; immunohistochemistry validation at https://www.stemcell.com/anti-beta-tubulin-iii-antibody-clone-tuj1.html and peer-reviewed citations at https://www.bioz.com/result/tuj1/product/STEMCELL%20Technologies%20Inc/?r=3.00&cf=0&uq=Stemcell%20Tech%20tuj1). Secondary antibodies were Alexa Fluor 488 Goat anti-Rabbit IgG (Thermo Fisher Scientific, Cat. # A-11008; 1:1,000; immunohistochemistry validation and peer-reviewed citations at https://www.thermofisher.com/antibody/product/Goat-anti-Rabbit-IgG-H-L-Cross-Adsorbed-Secondary-Antibody-Polyclonal/A-11008), Alexa Fluor 488 Goat anti-Mouse IgG (Thermo Fisher Scientific, Cat. # A-11001; 1:1,000; immunohistochemistry validation and peer-reviewed citations at https://www.thermofisher.com/antibody/product/Goat-anti-Mouse-IgG-H-L-Cross-Adsorbed-Secondary-Antibody-Polyclonal/A-11001), Texas Red-X Goat anti-Rat IgG (Thermo Fisher Scientific, Cat. # T-6392; 1:1,000; immunohistochemistry validation and peer-reviewed citations at https://www.thermofisher.com/antibody/product/Goat-anti-Rat-IgG-H-L-Cross-Adsorbed-Secondary-Antibody-Polyclonal/T-6392). DAPI (Thermo Fisher Scientific, Cat. # 62248; 1:1,000) was used as nuclear counterstaining and slides were mounted in FluroSave reagent (Millipore, Cat. # 345789).
For BrdU staining, sections were treated with 2 N HCl at 37 °C for 30 min and 0.1 M borate buffer, pH 8.5, for 10 min at room temperature.
Pax6-Tbr2 and Pax6-Mettl14 co-staining were performed according to the protocol described in a previous paper54. In brief, sections were incubated with highly diluted (1:30,000) primary antibody overnight at 4 °C and biotinylated goat anti-rabbit secondary antibody for 1 h at room temperature. The signal was amplified with the horseradish-peroxidase-based Vectastain ABC Kit (Vector Laboratories, Cat. # PK-6101) and Cyanine 3 Tyramide System (Perkin Elmer, Cat. # NEL704A001KT).
Fluorescence images were acquired by Zeiss LSM 710 confocal microscope and analyzed in ImageJ software.
NPC isolation and culture
Cortex region was dissected out from embryonic brains and triturated by pipetting. Dissociated cells were cultured as neurospheres with NeuroCult Proleferation Medium (Stemcell Tech.) according to the manufacturer’s protocols. Lentiviral constructs harboring shRNAs against Alkbh5 or Fto were purchased from Sigma-Aldrich (see below for details). Stable knockdown lines were generated using standard viral infection and puromycin selection (2 μg/ml).
shRNA RNA sequences
shAlkbh5-1: 5′-CCTATGAGTCCTCGGAAGATT-3′
shAlkbh5-1: 5′-GATCCTGGAAATGGACAAAGA-3′
shFto-1: 5′-GTCTCGTTGAAATCCTTTGAT-3′
shFto-1: 5′-TAGTCTGACTTGGTGTTAAAT-3′
Purification of mRNA
Total RNA was isolated using TRIZOL (Invitrogen) and treated with DNase I (Roche). Polyadenylated mRNA was purified using the GenElute mRNA Miniprep Kit (Sigma-Aldrich), and residual ribosomal RNA was depleted with RiboMinus Eukaryote System v2 (Life Technologies).
2D-TLC
2D-TLC was performed similarly as described55. Briefly, 500 ng purified polyA+/ribo– RNA was digested with 20 U RNase T1 (Thermo Fisher Scientific) in 20 µl 1× PNK buffer for 2 h at 37 °C. The digested RNA was labeled using 10 U T4 PNK (Thermo Fisher Scientific) and 1 µl γ-32P-ATP (6,000 Ci/mmol, 150 mCi/ml, Perkin-Elmer) and for 1 h at 37 °C and precipitated. The RNA pellet was resuspended in 5 µl buffer containing 25 mM NaCl, 2.5 mM ZnSO4 and 1 U nuclease P1 (Sigma) and incubated at 37 °C overnight. 1 µl product was loaded onto a PEI-Cellulose TLC plate (Millipore) and developed in isobutyric acid: 0.5 M NH4OH (5:3, v/v) as first dimension and isopropanol:HCl:water (70:15:15, v/v/v) as second dimension. After development, the plate was exposed to a phosphor screen and scanned using a FujiFilm FLA-5100 imager.
m6A dot-blot
Purified polyA+ RNA was blotted to a nylon membrane (Millipore) using Bio-Dot Microfiltration Apparatus (Bio-Rad). After cross-linking with a UV cross-linker (Spectroline), the membrane was blocked with nonfat milk in TBST and then incubated with antibody to m6A (Synaptic Systems, Cat. # 202 003, 1:1,000; dot blot validation and peer-reviewed citations at https://www.sysy.com/products/m6a/facts-202003.php) then an HRP-conjugated antibody to rabbit IgG (SouthernBiotech, Cat # 4030-05, 1:5,000; immunoblotting validation and peer-reviewed citations at https://www.southernbiotech.com/?catno=4030-05&type=Polyclonal#&panel1-1). After incubation with the Immobilon Western Chemiluminescent HRP Substrate (Millipore), the membrane was exposed to autoradiography film (Kodak).
Neurosphere formation assay
Dissociated NPCs were seeded into a 96-well plate at 400 cells per well and cultured for 7 d, then the 96-well plate was scanned using a Celigo Imaging Cytometer (Nexcelom Bioscience) and the number and size of neurospheres were measured.
Immunostaing of cultured NPCs and TUNEL assay
NPCs cultured for 7 d in vitro were dissociated and plated into chamber slides. The slides were stained with antibody to Tuj1 (1:1,000; Stemcell Tech., Cat. # 01409). TUNEL assay was performed using the Click-iT Plus kit (Thermo Fisher Scientific) according to the manufacturer’s protocols.
Apoptosis FACS
Dissociated NPCs were incubated with the Dead Cell Apoptosis Kit with Annexin V Alexa Fluor 488 and propidium iodide (Thermo Fisher Scientific) according to the manufacturer’s protocols and analyzed by LSRFortessa Cell Analyzer (BD Biosciences).
MTT assay
Dissociated NPCs were seeded into a 96-well plate. Cell numbers were measured using the Cell Growth Determination Kit (Sigma) according to the manufacturer’s protocols at different time points.
Western blot analysis
Proteins were separated on SDS-PAGE gel, blotted onto PVDF membrane and detected with primary antibodies to Gapdh (Cell Signaling, Cat. # 5174, 1:20,000; western blot validation and peer-reviewed citations at https://www.cellsignal.com/products/primary-antibodies/gapdh-d16h11-xp-rabbit-mab/5174), Mettl14 (Sigma-Aldrich, Cat. # HPA038002, 1:5,000; western blot validation and peer-reviewed citations at https://www.sigmaaldrich.com/catalog/product/sigma/hpa038002?lang=en®ion=US), Flag (Sigma-Aldrich, Cat. # 1804, 1:10,000; western blot validation and peer-reviewed citations at https://www.sigmaaldrich.com/catalog/product/sigma/f1804?lang=en®ion=US), Alkhb5 (Sigma-Aldrich, Cat. # HPA007196, 1:2,000; western blot validation and peer-reviewed citations at https://www.sigmaaldrich.com/catalog/product/sigma/hpa007196?lang=en®ion=US) and Fto (PhosphoSolutions, Cat. # 597-FTO, 1:1,000; western blot validation and peer-reviewed citations at https://www.phosphosolutions.com/shop/fto-antibody/). Secondary antibodies were Goat Anti-Rabbit IgG-HRP (SouthernBiotech, Cat. # 4030-05; 1:10,000; western blot validation and peer-reviewed citations at https://www.southernbiotech.com/?catno=4030-05&type=Polyclonal#&panel1-1) and Rabbit Anti-Mouse IgG-HRP (SouthernBiotech, Cat. # 6170-05; 1:10,000; western blot validation and peer-reviewed citations at https://www.southernbiotech.com/?catno=6170-05&type=Polyclonal).
Histone extraction and quantitative western blot analysis
Histones were extracted from NPCs cultured for 7 d in vitro using a Histone Extraction Kit (Abcam) according to the manufacturer’s protocols. Histone lysates were separated on SDS-PAGE gel, blotted onto Immobilon-FL PVDF membrane (Millipore, Cat. # IPFL00010) and incubated with primary antibodies including rat anti-H3 (Active Motif, Cat. # 61647; 1:2000; western blot validation and peer-reviewed citations at https://www.activemotif.com/catalog/details/61647/histone-h3-antibody-mab-clone-1c8b2), rabbit anti-H3K4me1 (Abcam, Cat. # Ab8895; 1:4,000; western blot validation and peer-reviewed citations at http://www.abcam.com/histone-h3-mono-methyl-k4-antibody-chip-grade-ab8895.html), rabbit anti-H3K4me3 (Abcam, Cat. # Ab8580; 1:2,000; western blot validation and peer-reviewed citations at http://www.abcam.com/histone-h3-tri-methyl-k4-antibody-chip-grade-ab8580.html), rabbit anti-H3K9me3 (Abcam, Cat. # Ab8898; 1:2,000; western blot validation and peer-reviewed citations at http://www.abcam.com/histone-h3-tri-methyl-k9-antibody-chip-grade-ab8898.html), rabbit anti-H3K27me3 (Millipore, Cat. # 07-449; 1:2,000; western blot validation at http://www.emdmillipore.com/US/en/product/Anti-trimethyl-Histone-H3-Lys27-Antibody,MM_NF-07-449 and peer-reviewed citations at https://www.bioz.com/result/antih3k27me3/product/Millipore/?r=4.88&cf=0&uq=Millipore, Cat. - 07%252D449), rabbit anti-H3K9ac (Active Motif, Cat. # 39917; 1:2,000; western blot validation at http://www.activemotif.com/catalog/details/39917 and peer-reviewed citations at https://www.bioz.com/result/h3k9ac/product/Active%20Motif/?r=4.14&cf=0&uq=Active%20Motif, Cat. - 39917), rabbit anti-H3K27ac (Abcam, Cat. # Ab4729; 1:2000; western blot validation and peer-reviewed citations at http://www.abcam.com/histone-h3-acetyl-k27-antibody-chip-grade-ab4729.html), and rabbit anti-H3ac (Active Motif, Cat. # 39139; 1:2,000; western blot validation at http://www.activemotif.com/catalog/details/39139/histone-h3ac-pan-acetyl-antibody-pab-1 and peer-reviewed citations at https://www.bioz.com/result/chip%20validated%20antibodies%20h3%20pan%20acetyl/product/Active%20Motif/?r=3.00&cf=0&uq=Active%20Motif, Cat. - 39139), rabbit anti-H2AK119Ubi (Cell Signaling, Cat. # 8240; 1:4000; western blot validation and peer-reviewed citations at https://www.cellsignal.com/products/primary-antibodies/ubiquityl-histone-h2a-lys119-d27c4-xp-rabbit-mab/8240), mouse anti-H2BK120Ubi (Active Motif, Cat. # 39623; 1:2000; western blot validation at http://www.activemotif.com/catalog/details/39623/histone-h2b-ubiquityl-lys120-antibody-mab-clone-56 and peer-reviewed citations at https://www.bioz.com/result/h2bub1/product/Active%20Motif/?r=3.00&cf=0&uq=Active%20Motif Motif/?r=3.00&cf=0&uq=Active Motif, Cat. - 39623), rabbit anti-H3S28pho (Abcam, Cat. # Ab10543; 1:2000; western blot validation at http://www.abcam.com/histone-h3-phospho-s28-antibody-hta28-ab10543.html and peer-reviewed citations at https://www.bioz.com/result/ab10543/product/Abcam/?r=3.00&cf=0&uq=Abcam, Cat. - Ab10543). Secondary antibodies were IRDye 800CW Goat anti-Rat IgG (LI-COR Biosciences, Cat. # 925-32219; 1:20,000; western blot validation and peer-reviewed citations at https://www.bioz.com/result/irdye%20800%20goat%20anti%20rat%20igg/product/LI-COR/?r=3.42&cf=0&uq=IRDye%20800%20Goat%20anti-Rat%20IgG), IRDye 680RD Goat anti-Rabbit IgG (LI-COR Biosciences, Cat. # 925-68071; 1:20,000; western blot validation and peer-reviewed citations at https://www.bioz.com/result/irdye%20680rd%20conjugated%20goat%20anti%20rabbit%20igg%20secondary%20antibody/product/LI-COR/?r=3.53&cf=0&uq=IRDye%20680RD%20Goat%20anti-Rabbit%20IgG), IRDye 680RD Goat anti-Mouse IgG (LI-COR Biosciences, Cat. # 925-68070; 1:20,000; western blot validation and peer-reviewed citations at https://www.bioz.com/result/irdye%20680rd%20goat%20anti%20mouse%20igg/product/LI-COR/?r=4.95&cf=0&uq=IRDye%20680RD%20Goat%20anti-Mouse%20IgG). Membranes were scanned by Odyssey Imager (LI-COR Biosciences) and quantified using Image Studio Lite (LI-COR Biosciences).
RT-qPCR
DNase-I-treated total RNA was used to synthesize cDNA using an iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer’s protocols. qPCR was performed with primer sets corresponding to the primer list table (Supplementary Table 7) and using iTaq Universal SYBR Green Supermix (Bio-Rad) on a BioRad CFX96 Touch Real-Time PCR Detection.
RNA-seq analysis
RNA-seq alignment
Tuxedo-suite was used for the entire RNA-seq analyses56. Specifically, TopHat2 (v2.1.0) was used to align RNA-seq reads to the mouse genome (mm10). To ensure correct alignment especially for assigning the spliced reads to the correct transcripts, we supplied the software with an existing transcript reference annotation from RefSeq with option–GTF. The RefSeq annotation is available as the iGenome project at the TopHat website (https://ccb.jhu.edu/software/tophat/igenomes.shtml). Other settings of the aligner were left to the default.
Transcript assembly
We then supplied the alignment BAM files from TopHat2 to Cufflinks to perform transcript assembly and expression estimation. The assembly was guided by the above RefSeq annotations with option–GTF-guide. We also turn on the multi-read correction (--multi-read-correct) and fragment bias correction options (--frag-bias-correct $genome) to correct for the sequencing biases. Other settings of the assembler were left to the default.
Differential analysis
We assembled transcripts in each sample library and biological replicate separately. In the end, we ran Cuffmerge to combine all assembled transcripts into a merged custom annotation file (merged.gtf) for the differential expression analysis. In particular, we ran Cuffdiff to compare between M14 knockout mutant and wild-type libraries using the read alignment files from TopHat2 as input and the above custom annotation file as reference. The 3 biological replicates per condition were used to estimate the dispersion in the negative binomial likelihood function modeled by Cuffdiff. We also enabled --multi-read-correct and --frag-bias-correct to improve estimation. The main output from Cuffdiff that were used for downstream analysis in this paper were the “gene_exp.diff” and “genes.read_group_tracking” (default output names from the software), which provide the differential expression statistical results and the actual expression estimate in terms of FPKM, respectively. Significant fold-change was determined based on FDR < 0.05 (as the default threshold of the Cuffdiff program).
GO analysis
GO analyses were performed by DAVID. Differentially expressed genes had an adjusted P < 0.01 and a twofold or greater expression difference.
ChIP-seq analysis
Chromatin immunoprecipitation (ChIP)
For ChIP experiments, NPCs cultured for 7 d in vitro were harvested and cross-linked for 5 min at room temperature using 1% formaldehyde. Cells were lysed by rotation for 10 min at 4 °C in cell lysis buffer (20 mM Tris, pH 8, 85 mM KCl, 0.5% NP-40, protease inhibitor cocktail). Nuclei were pelleted and re-suspended in nuclei lysis buffer (50 mM Tris, pH 8, 10 mM EDTA, 0.25% SDS, protease inhibitor cocktail). Chromatin was sheared by Covaris S2 using AFA microtube and a low cell chromatin shearing protocol for 12 min. Lysates were cleared by centrifugation at 20,000_g_ at 4 °C for 5 min and used as IP inputs. IP was done using the MAGnify Chromatin Immunoprecipitation System (Thermo Fisher Scientific) according to the manufacturer’s protocols. The antibodies used were as follows: rabbit anti-H3K27ac (Abcam, Cat. # Ab4729, Lot. # GR312658-1; ChIP validation and peer-reviewed citations at http://www.abcam.com/histone-h3-acetyl-k27-antibody-chip-grade-ab4729.html), rabbit anti-H3K27me3 (Millipore, Cat. # 07-449, Lot. # 2686928; ChIP validation at http://www.emdmillipore.com/US/en/product/Anti-trimethyl-Histone-H3-Lys27-Antibody,MM_NF-07-449 and peer-reviewed citations at https://www.bioz.com/result/antih3k27me3/product/Millipore/?r=4.88&cf=0&uq=Millipore, Cat. - 07%252D449). Chromatin samples were sent for high-throughput sequencing.
ChIP-seq alignment, peak calling, and comparison
To align ChIP-seq DNA reads to the reference mm10 genome, we used Bowtie2 (v2.3.1)57 with the default options except that “--broad” was turned on for calling broad peaks. Each ChIP library in each biological replicate was compared against the DNA input background library in the corresponding cell type + condition. Specifically, histone marks H3K4me3 and H3K27ac in wild-type and M14 KO mutant of mouse neural progenitor cell (NPC) were compared with the DNA input in wild-type and M14 KO NPC, respectively. Default cutoffs of FDR < 0.05 were used to determine ChIP-seq peaks relative to the input libraries. The main outputs from MACS2 were the broadPeak files in BED format, which we used for downstream comparison among the experimental conditions and with other cognate genome-wide results (that is, RNA-seq and RIP-seq).
meRIP-seq analysis
meRIP
meRIP was performed as previously described4. Briefly, total RNA was extracted using Trizol reagent (Invitrogen). RNA was treated with RNase-free DNase I (Roche) to deplete DNA contamination. PolyA RNA was purified using a GenElute mRNA Miniprep Kit (Sigma-Aldrich) as per the manufacturer’s instructions and fragmented using an RNA fragmentation kit (Ambion). 200 µg of fragmented RNA was incubated with 3 μg anti-m6A (Synaptic Systems, catalog number 202 003; RIP validation and peer-reviewed citations at https://www.sysy.com/products/m6a/facts-202003.php) in RIP buffer (150 mM NaCl, 10 mM Tris and 0.1% NP-40) for 2 h at 4 °C, followed by the addition of washed protein A/G magnetic beads (Millipore) and incubation at 4 °C for a further 2 h. Beads were washed 6 times in RIP buffer and incubated with 50 μl immunoprecipitation buffer containing 0.5 mg ml−1 m6AMP (Sigma-Aldrich) to elute RNA. Immunoprecipitated RNA was extracted with phenol/chloroform, and RNA samples were sent for high-throughput sequencing.
meRIP-seq alignment and peak calling
We used Tophat2 to align reads to the mouse reference mm10 genome with default setting and RefSeq annotations as a guide (that is, the same method as in the RNA-seq analysis). To call RIP-seq peaks, we used MACS2 with default settings except for enabling the --broad option (that is, the same as in the ChIP-seq analysis).
RNA stability assay
Actinomycin D (Sigma-Aldrich) at 5 μg/ml was added to NPC culture. After 0, 3 or 6 h of incubation, cells were collected and RNAs were isolated for qPCR.
Statistical analysis
All data are expressed as mean ± s.d., as indicated in the figure legends. Statistical analyses were completed with Prism GraphPad 7. Data comparing WT versus Mettl14 Het and KO phenotypes and data of inhibitor treatment were analyzed using one-way ANOVA followed by Bonferroni’s multiple comparisons test. Data comparing WT versus _Mettl14_-KO were analyzed using unpaired _t_-test. NSC growth curves were analyzed using two-way ANOVA followed by Bonferroni’s multiple comparisons test. RNA stability data comparing WT versus _Mettl14_-KO NSCs were analyzed using two-way ANOVA. The association between m6A targets and differential gene expression in _Mettl14_-KO versus nondeleted control NSCs were determined by Fisher’s exact test. Correlation analyses between changes in ChIP peak intensity and changes in gene expression were done using Pearson correlation analysis. No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications1,2,4,13,16,18. Data distribution was assumed to be normal but this was not formally tested. There was no randomization in the organization of the experimental conditions. Data collection and analysis were not performed blind to the conditions of the experiments. We did not exclude any animals for data point from the analysis.
Accession codes
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE104686.
Life Sciences Reporting Summary
Further information on experimental design is available in the Life Sciences Reporting Summary.
Data availability
RNA-seq and ChIP-seq data are accessible at NCBI GEO: GSE104686. The remaining data that support the findings of this study are available from the corresponding author upon reasonable request.
Change history
07 June 2018
In the version of this article initially published online, there were errors in URLs for www.southernbiotech.com, appearing in Methods sections “m6A dot-blot” and “Western blot analysis.” The first two URLs should be https://www.southernbiotech.com/?catno=4030-05&type=Polyclonal#&panel1-1 and the third should be https://www.southernbiotech.com/?catno=6170-05&type=Polyclonal. In addition, some Methods URLs for bioz.com, www.abcam.com and www.sysy.com were printed correctly but not properly linked. The errors have been corrected in the PDF and HTML versions of this article.
References
- Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012).
Article PubMed PubMed Central CAS Google Scholar - Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Article PubMed CAS Google Scholar - Liu, J. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95 (2014).
Article PubMed CAS Google Scholar - Wang, Y. et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16, 191–198 (2014).
Article PubMed PubMed Central CAS Google Scholar - Sledz, P. & Jinek, M. Structural insights into the molecular mechanism of the m(6)A writer complex. eLife 5, e18434 (2016).
Article PubMed PubMed Central Google Scholar - Wang, X. et al. Structural basis of N-adenosine methylation by the METTL3-METTL14 complex. Nature 534, 575–578 (2016).
Article PubMed CAS Google Scholar - Wang, P., Doxtader, K. A. & Nam, Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 63, 306–317 (2016).
Article PubMed PubMed Central CAS Google Scholar - Wang, Y. & Zhao, J. C. Update: mechanisms underlying N6-methyladenosine modification of eukaryotic mRNA. Trends Genet. 32, 763–773 (2016).
Article PubMed PubMed Central CAS Google Scholar - Jia, G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011).
Article PubMed PubMed Central CAS Google Scholar - Çağlayan, A. O. et al. A patient with a novel homozygous missense mutation in FTO and concomitant nonsense mutation in CETP. J. Hum. Genet. 61, 395–403 (2016).
Article PubMed PubMed Central CAS Google Scholar - Boissel, S. et al. Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. Am. J. Hum. Genet. 85, 106–111 (2009).
Article PubMed PubMed Central CAS Google Scholar - Hu, W. F., Chahrour, M. H. & Walsh, C. A. The diverse genetic landscape of neurodevelopmental disorders. Annu. Rev. Genomics Hum. Genet. 15, 195–213 (2014).
Article PubMed CAS Google Scholar - Geula, S. et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347, 1002–1006 (2015).
Article PubMed CAS Google Scholar - Hardwick, L. J. & Philpott, A. Nervous decision-making: to divide or differentiate. Trends Genet. 30, 254–261 (2014).
Article PubMed PubMed Central CAS Google Scholar - Zheng, G. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 (2013).
Article PubMed CAS Google Scholar - Hess, M. E. et al. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci. 16, 1042–1048 (2013).
Article PubMed CAS Google Scholar - Mauer, J. et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 541, 371–375 (2017).
Article PubMed CAS Google Scholar - Martynoga, B., Morrison, H., Price, D. J. & Mason, J. O. Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev. Biol. 283, 113–127 (2005).
Article PubMed CAS Google Scholar - Batista, P. J. et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719 (2014).
Article PubMed PubMed Central CAS Google Scholar - Wang, X. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014).
Article PubMed CAS Google Scholar - Bannister, A. J. & Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 (2011).
Article PubMed PubMed Central CAS Google Scholar - Avgustinova, A. & Benitah, S. A. Epigenetic control of adult stem cell function. Nat. Rev. Mol. Cell Biol. 17, 643–658 (2016).
Article PubMed CAS Google Scholar - Castelo-Branco, G. et al. Neural stem cell differentiation is dictated by distinct actions of nuclear receptor corepressors and histone deacetylases. Stem Cell Rep. 3, 502–515 (2014).
Article PubMed PubMed Central CAS Google Scholar - Qiao, Y., Wang, R., Yang, X., Tang, K. & Jing, N. Dual roles of histone H3 lysine 9 acetylation in human embryonic stem cell pluripotency and neural differentiation. J. Biol. Chem. 290, 2508–2520 (2015).
Article PubMed CAS Google Scholar - Román-Trufero, M. et al. Maintenance of undifferentiated state and self-renewal of embryonic neural stem cells by Polycomb protein Ring1B. Stem Cells 27, 1559–1570 (2009).
Article PubMed CAS Google Scholar - Voncken, J. W. et al. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc. Natl. Acad. Sci. USA 100, 2468–2473 (2003).
Article PubMed PubMed Central CAS Google Scholar - cIshino, Y. et al. Bre1a, a histone H2B ubiquitin ligase, regulates the cell cycle and differentiation of neural precursor cells. J. Neurosci. 34, 3067–3078 (2014).
Article CAS Google Scholar - Pereira, J. D. et al. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc. Natl. Acad. Sci. USA 107, 15957–15962 (2010).
Article PubMed PubMed Central Google Scholar - Mohn, F. et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell 30, 755–766 (2008).
Article PubMed CAS Google Scholar - Nakashima, K. et al. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science 284, 479–482 (1999).
Article PubMed CAS Google Scholar - Wang, J. et al. CBP histone acetyltransferase activity regulates embryonic neural differentiation in the normal and Rubinstein-Taybi syndrome brain. Dev. Cell 18, 114–125 (2010).
Article PubMed CAS Google Scholar - Lim, D. A. et al. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature 458, 529–533 (2009).
Article PubMed PubMed Central CAS Google Scholar - Roopra, A., Qazi, R., Schoenike, B., Daley, T. J. & Morrison, J. F. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol. Cell 14, 727–738 (2004).
Article PubMed CAS Google Scholar - Boyer, L. A. et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353 (2006).
Article PubMed CAS Google Scholar - Bernstein, B. E. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006).
Article PubMed CAS Google Scholar - Creyghton, M. P. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 107, 21931–21936 (2010).
Article PubMed PubMed Central Google Scholar - Wang, Z. et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 40, 897–903 (2008).
Article PubMed PubMed Central CAS Google Scholar - Zhou, R., Niwa, S., Homma, N., Takei, Y. & Hirokawa, N. KIF26A is an unconventional kinesin and regulates GDNF-Ret signaling in enteric neuronal development. Cell 139, 802–813 (2009).
Article PubMed CAS Google Scholar - Ju, Y. T. et al. gas7: A gene expressed preferentially in growth-arrested fibroblasts and terminally differentiated Purkinje neurons affects neurite formation. Proc. Natl. Acad. Sci. USA 95, 11423–11428 (1998).
Article PubMed PubMed Central CAS Google Scholar - Williams, B. P. et al. A PDGF-regulated immediate early gene response initiates neuronal differentiation in ventricular zone progenitor cells. Neuron 18, 553–562 (1997).
Article PubMed CAS Google Scholar - Li, S. et al. The transcription factors Egr2 and Egr3 are essential for the control of inflammation and antigen-induced proliferation of B and T cells. Immunity 37, 685–696 (2012).
Article PubMed PubMed Central CAS Google Scholar - Alarcón, C. R. et al. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell 162, 1299–1308 (2015).
Article PubMed PubMed Central CAS Google Scholar - Xiao, W. et al. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61, 507–519 (2016).
Article PubMed CAS Google Scholar - Ben-Haim, M. S., Moshitch-Moshkovitz, S. & Rechavi, G. FTO: linking m6A demethylation to adipogenesis. Cell Res. 25, 3–4 (2015).
Article PubMed CAS Google Scholar - Meyer, K. D. et al. 5′ UTR m(6)A promotes cap-independent translation. Cell 163, 999–1010 (2015).
Article PubMed PubMed Central CAS Google Scholar - Wang, X. et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399 (2015).
Article PubMed PubMed Central CAS Google Scholar - Greer, E. L. & Shi, Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 13, 343–357 (2012).
Article PubMed PubMed Central CAS Google Scholar - Karatas, H. et al. High-affinity, small-molecule peptidomimetic inhibitors of MLL1/WDR5 protein-protein interaction. J. Am. Chem. Soc. 135, 669–682 (2013).
Article PubMed CAS Google Scholar - Lee, E. C. et al. A highly efficient _Escherichia coli_-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73, 56–65 (2001).
Article PubMed CAS Google Scholar - Ogawa, Y. & Lee, J. T. Xite, X-inactivation intergenic transcription elements that regulate the probability of choice. Mol. Cell 11, 731–743 (2003).
Article PubMed CAS Google Scholar - Yamada, N. et al. Xist exon 7 contributes to the stable localization of Xist RNA on the inactive X-chromosome. PLoS Genet. 11, e1005430 (2015).
Article PubMed PubMed Central CAS Google Scholar - Daikoku, T. et al. Lactoferrin-iCre: a new mouse line to study uterine epithelial gene function. Endocrinology 155, 2718–2724 (2014).
Article PubMed PubMed Central CAS Google Scholar - George, S. H. et al. Developmental and adult phenotyping directly from mutant embryonic stem cells. Proc. Natl. Acad. Sci. USA 104, 4455–4460 (2007).
Article PubMed PubMed Central CAS Google Scholar - Arai, Y. et al. Neural stem and progenitor cells shorten S-phase on commitment to neuron production. Nat. Commun. 2, 154 (2011).
Article PubMed PubMed Central CAS Google Scholar - Bodi, Z., Button, J. D., Grierson, D. & Fray, R. G. Yeast targets for mRNA methylation. Nucleic Acids Res. 38, 5327–5335 (2010).
Article PubMed PubMed Central CAS Google Scholar - Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Article PubMed PubMed Central CAS Google Scholar - Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Article PubMed PubMed Central CAS Google Scholar
Acknowledgements
We thank L. Wang and V. Ylis from the Sanford-Burnham-Prebys Medical Discovery Institute (SBP) Animal Resources Facility for blastocyst injection and mouse maintenance; G. Garcia from the SBP Histopathology core for preparing coronal sections from frozen and paraffin-embedded mouse brains; the SBP Cell Imaging core for confocal microscopy; the SBP flow cytometry core for apoptosis analysis; B. James from the Genomics DNA Analysis Facility at SBP for high-throughput sequencing analysis; and A. Bang for critical reading of the manuscript. This work is supported by a CIHR Operating grant (No. 115194) and an NSERC Discovery Grant (327612) to Z.Z., an NCI grant (CA159859) and a CIRM (California Institute for Regenerative Medicine) Leadership Award (LA1-01747) to R.W.R., an NIGMS grant (GM062848) to G.D., NIH R01 awards (MH109978 and HG008155), an NIH RF1 award (AG054012), and an NIH U01 award (HG007610) to M.K., a CIRM Training Grant (TG2-01162) to Y.W., an NIH R01 award (R01 GM110090) and an SBP Cancer Center Pilot grant (5P30 CA030199) to J.C.Z.
Author information
Authors and Affiliations
- Tumor Initiation and Maintenance Program, NCI-designated Cancer Center, Sanford Burnham Prebys Medical Discovery Institute, La Jolla, CA, USA
Yang Wang, Jun Wang, Robert J. Wechsler-Reya & Jing Crystal Zhao - Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, MA, USA
Yue Li & Manolis Kellis - Division of Reproductive Sciences, Division of Developmental Biology, Perinatal Institute, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA
Minghui Yue & Yuya Ogawa - Department of Pediatrics, University of Cincinnati, Cincinnati, OH, USA
Minghui Yue & Yuya Ogawa - Development, Aging, and Regeneration Program, Sanford Burnham Prebys Medical Discovery Institute, La Jolla, CA, USA
Sandeep Kumar & Gregg Duester - Department of Molecular Genetics, The Donnelly Centre, University of Toronto, Toronto, Ontario, Canada
Zhaolei Zhang
Authors
- Yang Wang
You can also search for this author inPubMed Google Scholar - Yue Li
You can also search for this author inPubMed Google Scholar - Minghui Yue
You can also search for this author inPubMed Google Scholar - Jun Wang
You can also search for this author inPubMed Google Scholar - Sandeep Kumar
You can also search for this author inPubMed Google Scholar - Robert J. Wechsler-Reya
You can also search for this author inPubMed Google Scholar - Zhaolei Zhang
You can also search for this author inPubMed Google Scholar - Yuya Ogawa
You can also search for this author inPubMed Google Scholar - Manolis Kellis
You can also search for this author inPubMed Google Scholar - Gregg Duester
You can also search for this author inPubMed Google Scholar - Jing Crystal Zhao
You can also search for this author inPubMed Google Scholar
Contributions
J.C.Z. and Y.W. formulated the idea. Y.W. designed, performed and analyzed most of the experiments. Y.L. performed bioinformatics analysis under the supervision of M.K. Z.Z. provided additional suggestions relevant to bioinformatics analysis. M.Y. and Y.O. generated floxed Mettl14 mESCs for blastocyst injection. J.W. and R.W.R. provided technical help on mouse brain sectioning and NSC culture, and helped with data analysis. S.K. and G.D. helped with dissection of early embryos. J.C.Z. and Y.W. analyzed data and wrote the paper.
Corresponding author
Correspondence toJing Crystal Zhao.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations
Integrated supplementary information
Supplementary Figure 1 Generation of conditional _Mettl14_-knockout mice
(a) Schematic diagram showing strategy used to create conditional Mettl14 knockout. (b-d) Representative genotyping PCR results using different primer sets for the targeted Mettl14 allele (b), the floxed Mettl14 allele (c), or the exon 2-deleted Mettl14 allele (d). Similar results were obtained from three independent experiments. (e) Representative images of wildtype and Mettl14 null embryos at E7.5. Similar results were obtained from three independent experiments. (f) Representative PCR results for sex determination. Similar results were obtained from three independent experiments. (g) Representative images of pups at P0. Scale bar: 5 mM. (h) Quantification of pup body weight at P0, one-way ANOVA (WT: n = 16, KO: n = 7, and Het: n = 8 P0 pups; P = 0.1719, F (2, 28) = 1.877) followed by Bonferroni's post hoc test (WT vs. KO, P = 0.4198, 95% C.I. = −0.09762 to 0.3285, WT vs. Het, P = 0.6836, 95% C.I. = −0.2867 to 0.1205). (i) Survival curve of newborn mice with or without brain-specific Mettl14 deletion (Mettl14 f/f: n = 12, Mettl14 f/+: n = 10, Mettl14 f/+;nes-cre: n = 12, Mettl14 f/f;nes-cre: n = 14 animals). Graphs represent mean ± SD. Dots represent data from individual data points. ns = non-significant.
Supplementary Figure 2 Mettl14 regulates self-renewal of cortical NSCs in vitro
(a) Western blots showing Mettl14 depletion in Mettl14 KO NSCs. Similar results were obtained from three independent experiments. For uncropped images, see Supplementary Fig. 6b. (b) Growth curve of cortical NSCs isolated from E17.5 brain. Two-way ANOVA (n = 3 cell cultures for all experimental groups; P = 3E-15, F (2, 18) = 357.5) followed by Bonferroni's post hoc test (WT vs. KO, P = 2.5E-14, 95% C.I. = 0.09616 to 0.1196, WT vs. Het, P = 0.4346, 95% C.I. = −0.01786 to 0.005595). (c,d) Assessment of apoptosis in Mettl14 KO and nondeleted control NSCs. The number of apoptotic cells was determined by FACS analysis via Annexin V-FITC and PI-staining. Representative results are shown in (c) and results from 3 independent experiments are summarized in (d). One-way ANOVA (n = 3 cell cultures for all experimental groups; P = 0.6882, F (2, 6) = 0.3979) followed by Bonferroni's post hoc test (WT vs. KO, P = 0.8321, 95% C.I. = −1.999 to 3.666, WT vs. Het, P = 0.9999, 95% C.I. = −2.266 to 3.399). (e) Quantification of TUNEL assays in Mettl14 KO and control NSCs. One-way ANOVA (n = 3 fields for all experimental groups; P = 0.7572, F (2, 6) = 0.2915) followed by Bonferroni's post hoc test (WT vs. KO, P = 0.9999, 95% C.I. = −50.16 to 32.35, WT vs. Het, P = 0.9999, 95% C.I. = −40.71 to 41.81). (f) Western blots showing expression of Flag-tagged Mettl14 in WT and Mettl14 KO NSCs transduced by lentivirus containing empty or Flag-Mettl14 vectors. Similar results were obtained from three independent experiments. For uncropped images, see Supplementary Fig. 6c. (g) m6A dot-blots of Ribo- polyA RNAs isolated from Mettl14 KO and nondeleted NSCs transduced with lentivirus containing empty or Flag-Mettl14 vectors. Similar results were obtained from three independent experiments. (h) RT-qPCR of Alkbh5 transcripts in NSCs expressing scramble (scr) shRNA or shRNAs against Alkbh5, one-way ANOVA (n = 3 independent experiments for all experimental groups; P = 7.511E-08, F (2, 6) = 708) followed by Bonferroni's post hoc test (Scr vs. shAlkbh5-1, P = 1.06113E-07, 95% C.I. = 0.8453 to 1.013, Scr vs. shAlkbh5-2, P = 1.164E-07, 95% C.I. = 0.831 to 0.9991). (i) Western blots showing Alkbh5 depletion in NSCs expressing scramble (scr) shRNA or shRNAs against Alkbh5. Similar results were obtained from two independent experiments. For uncropped images, see Supplementary Fig. 6d. (j) Growth curve of NSCs expressing scr shRNA or shRNAs against Alkbh5. Two-way ANOVA (n = 4 cell cultures for all experimental groups; P = 0.0626, F (2, 27) = 3.075) followed by Bonferroni's post hoc test (Scr vs. shAlkbh5-1, P = 0.0928, 95% C.I. = −0.2273 to 3.548, Scr vs. shAlkbh5-2, P = 0.0726, 95% C.I. = −0.1353 to 3.64). (k) RT-qPCR of Fto transcripts in NSCs expressing scramble (scr) shRNA or shRNAs against Fto, one-way ANOVA (n = 3 independent experiments for all experimental groups; P = 1.324E-06, F (2, 6) = 270.2) followed by Bonferroni's post hoc test (Scr vs. shFto-1, P = 2.396E-06, 95% C.I. = 0.6521 to 0.887, Scr vs. shFto-2, P = 1.629E-06, 95% C.I. = 0.6521 to 0.887). (l) Western blots showing Fto depletion in NSCs expressing scramble (scr) shRNA or shRNAs against Fto. Similar results were obtained from two independent experiments. For uncropped images, see Supplementary Fig. 6e. (m) Growth curve of NSCs expressing scr shRNA or shRNAs against Fto. Two-way ANOVA (n = 4 cell cultures for all experimental groups; P = 0.0005, F (2, 27) = 10.1) followed by Bonferroni's post hoc test (Scr vs. shFto-1, P = 0.0538, 95% C.I. = −1.604 to 0.01121, Scr vs. shFto, P = 0.0809, 95% C.I. = −0.07525 to 1.54). Graphs represent mean ± SD. Dots represent data from individual data points. ns = non-significant. **** P < 0.0001.
Supplementary Figure 3 Mettl14 deficiency decreases RGC proliferation in vivo
(a-c) Coronal sections of E15.5 brains stained with antibodies recognizing BrdU, PH3, and PAX6. Pregnant mothers received a BrdU pulse 30 min prior to embryo collection. (d) Quantification of immunostaining from E15.5 sections. Numbers of Pax6+, BrdU+ and PH3+ cells were determined and normalized to comparable sections from nondeleted mice, one-way ANOVA (n = 3 brain sections for all experimental groups; Pax6+, P = 0.5647, F (2, 6) = 0.6296; BrdU+, P = 0.0159, F (2, 6) = 8.942; PH3+, P = 0.016, F (2, 6) = 8.912) followed by Bonferroni's post hoc test (Pax6+, WT vs. KO, P = 0.8495, 95% C.I. = −0.342 to 0.6193, WT vs. Het, P = 0.9999, 95% C.I. = −0.5131 to 0.4483; BrdU+, WT vs. KO, P = 0.0442, 95% C.I. = 0.007399 to 0.4702, WT vs. Het, P = 0.7184, 95% C.I. = −0.3088 to 0.154; PH3+, WT vs. KO, P = 0.0134, 95% C.I. = 0.1228 to 0.7966, WT vs. Het, P = 0.7164, 95% C.I. = −0.224 to 0.4498). (e,f) Coronal sections of E13.5 brain stained with anti-Pax6, anti-BrdU, or anti-PH3 antibodies. Pregnant mothers were pulsed with BrdU 30 min prior to embryo collection. (g) Quantification of immunostaining from E13.5 sections. Numbers of Pax6+, BrdU+ and PH3+ cells were determined and normalized to comparable sections from nondeleted mice, one-way ANOVA (n = 3 brain sections for all experimental groups; Pax6+, P = 0.7735, F (2, 6) = 0.2682; BrdU+, P = 0.6595, F (2, 6) = 0.4466; PH3+, P = 0.9675, F (2, 6) = 0.03325) followed by Bonferroni's post hoc test (Pax6+, WT vs. KO, P = 0.9999, 95% C.I. = −0.364 to 0.403, WT vs. Het, P = 0.9999, 95% C.I. = −0.454 to 0.3131; BrdU+, WT vs. KO, P = 0.8695, 95% C.I. = −0.2011 to 0.359, WT vs. Het, P = 0.9999, 95% C.I. = −0.2765 to 0.2836; PH3+, WT vs. KO, P = 0.9999, 95% C.I. = −0.1804 to 0.2045, WT vs. Het, P = 0.9999, 95% C.I. = −0.1764 to 0.2086). (h) Immunostaining of coronal sections of E17.5 and E15.5 brain using Cleaved-Caspase-3 antibody. (i) Quantification of Cleaved-Caspase-3+ cell number, based on immunostaining. One-way ANOVA (n = 3 brain sections for all experimental groups; E17.5, P = 0.1834, F (2, 6) = 2.28; E15.5, P = 0.6596, F (2, 6) = 0.4463) followed by Bonferroni's post hoc test (E17.5, WT vs. KO, P = 0.5745, 95% C.I. = −11.83 to 27.16, WT vs. Het, P = 0.744, 95% C.I. = −25.83 to 13.16; E15.5, WT vs. KO, P = 0.9999, 95% C.I. = −36.44 to 32.44, WT vs. Het, P = 0.9991, 95% C.I. = −26.11 to 42.77). (j,k) Immunostaining of coronal sections of E15.5 brain with antibodies to the intermediate progenitor marker anti-Tbr2 and the proneural marker anti-Neurod2. Dashed white lines indicate border of VZ/SVZ area. Similar results were obtained from three independent experiments. Scale bars: 100 μM. Graphs represent mean ± SD. Dots represent data from individual data points. ns = non-significant. * P < 0.05.
Supplementary Figure 4 m6A regulates NSC gene expression through histone modifications
(a) Numbers of gene counts and exact P values for each GO term displayed in Fig. 5b and 5c. (b) The association between m6A targets and differential gene expression in Mettl14 KO vs. nondeleted control NSCs determined by two-tailed Fisher’s Exact Test. A transcript is defined as an m6A target when a m6A peak is detected from either one of the two biological replicates in m6A meRIP-seq experiment. The numbers of genes in each category and exact P value are shown in the graph. (c) MTT cell growth assay of NSCs treated with vehicle/DMSO or the MLL1 inhibitor MM-102, the CBP/P300 inhibitor C646, or the Ezh2 inhibitor GSK343. Shown is the absorbance ratio of heterozygous to non-deleted controls at each dose. One-way ANOVA (n = 3 independent experiments for all experimental groups; GSK343, P = 0.1352, F (3, 8) = 2.482; C646, P = 0.0528, F (3, 8) = 3.971; MM-102, P = 0.5055, F (3, 8) = 0.8478) followed by Bonferroni's post hoc test (GSK343, c vs. 1.25, P = 0.9999, 95% C.I. = −0.2624 to 0.1392, c vs. 2.5, P = 0.0932, 95% C.I. = −0.3746 to 0.02694, c vs. 5, P = 0.3732, 95% C.I. = −0.3151 to 0.08651; C646, c vs. 1.25, P = 0.3132, 95% C.I. = −0.1995 to 0.04874, c vs. 2.5, P = 0.4435, 95% C.I. = −0.0582 to 0.1901, c vs. 5, P = 0.9999, 95% C.I. = −0.1164 to 0.1319; MM-102, c vs. 0.0625, P = 0.9999, 95% C.I. = −0.1997 to 0.3001, c vs. 1.25, P = 0.9999, 95% C.I. = −0.3251 to 0.1747, c vs. 2.5, P = 0.9999, 95% C.I. = −0.2263 to 0.2734). Graphs represent the mean ± SD. Dots represent data from individual data points.
Supplementary Figure 5 Low enrichment of m6A on mRNAs encoding Ezh2, Suz12, and Eed m6A-meRIP-qPCR of Ezh2, Suz12, and Eed in Mettl14 KO vs.
control E14.5 NSCs. One-way ANOVA (n = 3 independent experiments for all experimental groups; Ezh2, P = 0.0812, F (2, 6) = 3.928; Suz12, P = 5.055E-06, F (2, 6) = 171.8; Eed, P = 0.00012, F (2, 6) = 57.32) followed by Bonferroni's post hoc test (Ezh2, WT vs. KO, P = 0.0739, 95% C.I. = −0.004323 to 0.08219, WT vs. Het, P = = 0.1679, 95% C.I. = −0.0131 to 0.07341; Suz12, WT vs. KO, P = 3.125E-05, 95% C.I. = 0.2091 to 0.3387, WT vs. Het, P = 0.0029, 95% C.I. = −0.1855 to −0.05595; Eed, WT vs. KO, P = 9.828E-05, 95% C.I. = 0.02144 to 0.03883, WT vs. Het, P = 0.0823, 95% C.I. = −0.001105 to 0.01628). Graphs represent the mean ± SD. Dots represent data from individual data points. ns = non-significant. **** P < 0.0001.
Supplementary Figure 6 Uncropped western blot images
(a) Corresponding uncropped images of western blots shown in Fig. 5d. (b) Corresponding uncropped images of western blots shown in Supplementary Fig. 2a. (c) Corresponding uncropped images of western blots shown in Supplementary Fig. 2f. (d) Corresponding uncropped images of western blots shown in Supplementary Fig. 2i. (e) Corresponding uncropped images of western blots shown in Supplementary Fig. 2l. The protein standards are depicted to the left of the images.
Supplementary information
Life Sciences Reporting Summary
Supplementary Tables 1, 2, and 7
Supplementary Table 1 Genotypes of born pups from Mettl14_+/−× Mettl14+/−_ crosses
Supplementary Table 2 Genotypes of E7.5, E8.5, and E9.5 embryos from Mettl14_+/−_ × Mettl14_+/−_ crosses
Supplementary Table 3 List of PCR primers used in this study
Supplementary Table 3
RNA-seq analysis of E14.5 _Mettl14_-KO NSC vs. nondeleted control from 3 independent experiments
Supplementary Table 4
Peaks detected from methylated RIP-seq experiments. Results from the biological replicates were listed in two separate spreadsheets.
Supplementary Table 5
H3K27ac ChIP-seq peaks located within 10 Kb up- or downstream of transcription start sites
Supplementary Table 6
H3K27me3 ChIP-seq peaks located within 2 Kb upstream of transcription start sites
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0), which permits use, duplication, adaptation, distribution, and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Wang, Y., Li, Y., Yue, M. et al. _N_6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications.Nat Neurosci 21, 195–206 (2018). https://doi.org/10.1038/s41593-017-0057-1
- Received: 04 July 2017
- Accepted: 04 December 2017
- Published: 15 January 2018
- Issue Date: February 2018
- DOI: https://doi.org/10.1038/s41593-017-0057-1