Sprouty2 inhibits BDNF-induced signaling and modulates neuronal differentiation and survival (original) (raw)
Main
The neurotrophin BDNF (brain-derived neurotrophic factor) is widely expressed in the mammalian central nervous system (CNS) and plays a key role in its development and homeostasis.1 At the cellular level, BDNF induces neuronal differentiation and neurite outgrowth,2 facilitates plasticity,3 and favors neuronal survival.4 BDNF binds to the tyrosine kinase receptor TrkB, which activates at least three signal transduction pathways: (i) the Ras/mitogen-activated protein (MAP) kinase (Ras/MAPK) pathway; (ii) the PI3-Kinase/Akt pathway; and (iii) the PLC_γ_ pathway.5 Activation of these pathways allows BDNF to regulate transcription6 and ultimately accounts for its role in neuronal differentiation and survival.2
The coordinated action of extracellular signals that lead to brain development and maintenance requires a tight spatial and temporal control, which is partly achieved by negative-feedback loops, involving the clearance of the receptor, the phosphorylation state of proteins, or the expression of counteracting factors.7 Over the past few years, members of the Sprouty (Spry) family of proteins have emerged as critical modulators of growth factor signaling (for recent reviews, see Mason et al.8 and Kim and Bar-Sagi9). The mouse and human genome each contain four spry genes encoding proteins of 32–34 kDa. Spry proteins are generally negative regulators of receptor tyrosine kinase (RTK) signaling. The activity of Spry is regulated at the transcriptional level, with growth factors inducing Spry expression, and post-translational level as growth factors can lead to phosphorylation of Spry on critical tyrosine residues.8, 9 Genetic studies confirmed that spry genes are part of a negative-feedback loop to limit the effects of specific growth factors such as fibroblast growth factor (FGF) and glial cell-derived neurotrophic factor (GDNF). Mammalian spry genes are expressed in highly restricted patterns in the embryo during development and in many adult tissues. They were shown to play important roles in the development of various organs, including lung,10 kidney,11, 12 tooth13 and testis.14 At the cellular level, overexpression of Spry proteins antagonizes proliferation, migration and differentiation in response to serum or growth factors in various cell lines.8, 9
So far, the contribution of spry genes to neuronal differentiation or survival has not been explored in mammals, although they exhibit a strikingly localized expression pattern in the developing brain.15, 16 In view of the essential function of growth factors and neurotrophins in neuronal differentiation and survival through their activation of RTK signaling, we investigated the possible role of Spry2, a prototypical member of the Spry family, in neurons of the CNS.
Results
Spry2 is expressed in cerebellar granule neurons in vivo and ex vivo
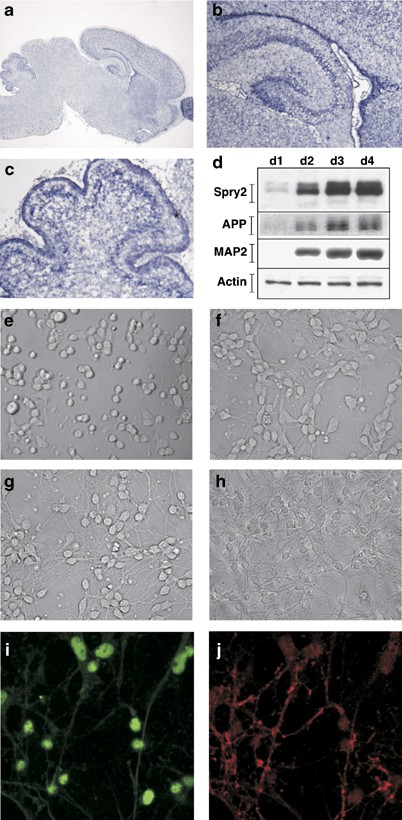
Primary cultures of mouse cerebellar granule neurons (CGN) are well suited to study neuronal differentiation and survival ex vivo, as they provide a homogeneous culture of neurons.17 To check whether this model would be appropriate to investigate the potential role and regulation of Spry2 in neural cells, we first examined spry2 expression in the brain of 5 days old mice by in situ hybridization (ISH). spry2 expression was detected in multiple areas of the brain: the cerebellum, the cortex, the hippocampus and the olfactory bulb (Figure 1a). Higher magnification observations revealed spry2 staining in the CA1, CA2 and CA3 (hippocampus, Figure 1b) and EGL (external granule cell layer, cerebellum, Figure 1c, Supplementary data S3). No signal was detected with a control probe (data not shown).
Figure 1
Expression of Spry2 in neurons. (a–c) Expression of spry2 in the developing brain. Cryostat sections (12 _μ_m) of a 5 days old mouse brain were subjected to ISH with a specific spry2 dig-labeled antisense RNA probe. (b) Shows a higher magnification of the hippocampus and (c) of the cerebellum. *Indicates localization of the EGL. (d) Western blot detection of Spry2 expression in primary CGN at the indicated day of culture. Cell extracts were analyzed by immunoblotting with antibodies directed against Spry2, MAP2, APP or Actin. (e–h) Morphology of CGN at day 1 to day 4 of the culture. (i, j) Immunocytofluorescence detection of Spry2 (i) and MAP2 (j) expression in differentiated CGN at day 4
Next, we checked Spry2 expression in a primary culture of mouse CGN. Neuron progenitors dissected from 7 days old mice were plated in a medium containing Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% horse serum and 25 mM KCl. At day 0, plated neurons were round cells without neurites (Figure 1e). From day 1 through day 4, these immature cells differentiated and grew neurites to form an active and complex neuronal network (Figure 1f–h).17 The neuronal differentiation status was confirmed by the progressive increase of neuronal markers expression, such as MAP2, amyloid β precursor protein (APP), NeuroD and beta-2 adrenergic receptor (Figure 1d and data not shown).18, 19, 20 At day 1, weak Spry2 expression was detected by Western blot (Figure 1d). This expression was stronger at day 2 and was further increased at days 3 and 4. Immunocytofluorescence performed with anti-Spry2 and anti-MAP2 antibodies at day 4 confirmed the co-expression of Spry2 and the neuronal marker MAP2 in the same cells that is, neurons (Figure 1i, j).
BDNF and serum regulate spry2 expression in cerebellar granule neurons
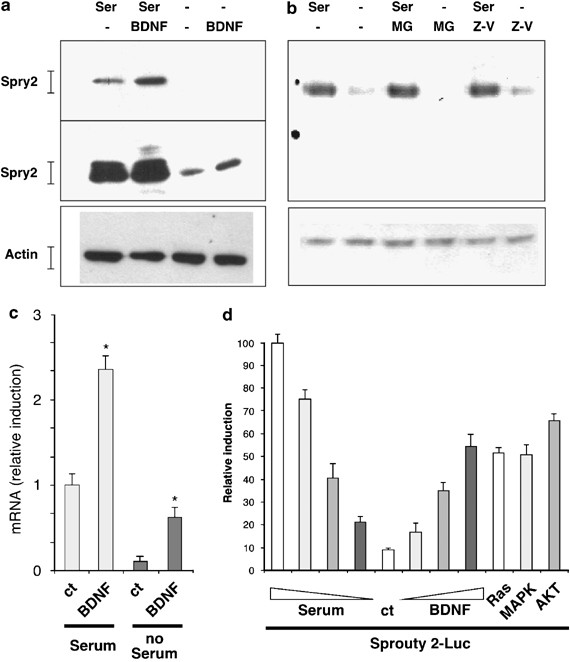
BDNF and other neurotrophins or growth factors (like FGF) have been implicated in the differentiation of cultured CGN through regulation of the Ras/MAPK pathway.2, 21 As spry expression is regulated by growth factors and is involved in a negative-feedback loop in various contexts,22, 23 we investigated whether BDNF regulated Spry2 levels in neurons. Expression of Spry2 was clearly detected by Western blot in neurons cultured in presence of serum. Culture of the neurons with BDNF further increased Spry2 expression (Figure 2a, upper panel). In the absence of serum, Spry2 protein levels were significantly lower, but could still be augmented by addition of BDNF to the culture medium (Figure 2a, medium panel).
Figure 2
Regulation of Spry2 expression by BDNF. (a) Effect of serum and BDNF on Spry2 protein levels in CGN. Immature CGN were grown in the indicated culture conditions for 16 h (Ser: serum 10%; BDNF: 25 ng/ml) before lysis. Cell extracts were analyzed by immunoblotting with an antibody against Spry2 (short exposure: top panel; long exposure: middle panel) and an antibody against Actin to control protein loading (bottom panel). (b) Effect of protease inhibitors on Spry2 protein levels: immature CGN were deprived from serum in presence of MG132 (MG; 25 _μ_M) or Z-VAD (25 _μ_M). At 18 h after treatment, cells were lysed and protein extracts were analyzed by immunoblotting with the indicated antibodies. (c) Serum and BDNF regulate spry2 mRNA levels in CGN. Neurons were grown for 16 h as indicated before lysis. Total RNA was submitted to reverse transcription, followed by real-time PCR amplification of spry2 and of 18s cDNA fragments. The graph shows spry2 relative expression after normalization with 18s levels and with spry2 expression in the presence of only serum set at 1. Bars represent means±S.D. in a representative experiment performed in triplicates. Asterisks indicate significant difference (P<0.05) compared to control, as calculated by a one-way ANOVA test, followed by a Newman–Keul's test. (d) BDNF and serum regulate the activity of the spry2 promoter. Immature CGN were transfected with a spry2 promoter-Luc reporter gene (−3850 luc). Cells were treated with different concentrations of BDNF (5, 15, 50 ng/ml) or of serum (0.1, 1, 10%) for 18 h. As indicated, cells were co-transfected with empty vector or with either Ras, Erk1 MAPK or Akt expression vectors. At 24 h after transfection, cells were lyzed and luc was quantified. Bars represent means±S.D. in a representative experiment performed in triplicates
Several studies have shown that the regulation of Spry2 protein levels by growth factor involved the proteasome.8, 24 Therefore, we tested the effect of a proteasome inhibitor on the disappearance of Spry2. Proteasome inhibitor or even a caspase inhibitor did not restore the protein levels of Spry2 (Figure 2b). On the other hand, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) showed that messenger RNA (mRNA) levels for spry2 were increased by the presence of serum and BDNF (Figure 2c). In the absence of serum, spry2 mRNA levels were reduced, whereas BDNF augmented spry2 mRNA levels. BDNF also augmented spry2 levels in the presence of serum. To corroborate these results, we tested the effects of serum and BDNF on the activity of the spry2 promoter.25 In accordance with the RNA data, removal of serum strongly attenuated spry2 promoter activity, whereas BDNF clearly increased it (Figure 2d). The transcriptional activity of the spry2 promoter was also stimulated by overexpression of intracellular relays activated by BDNF such as Ras, Erk1 MAPK, or Akt (Figure 2d). These results show that expression of spry2 in developing neurons is regulated at the transcriptional level by serum and BDNF.
BDNF regulates spry2 promoter activity through CREB and SP1 transcription factors
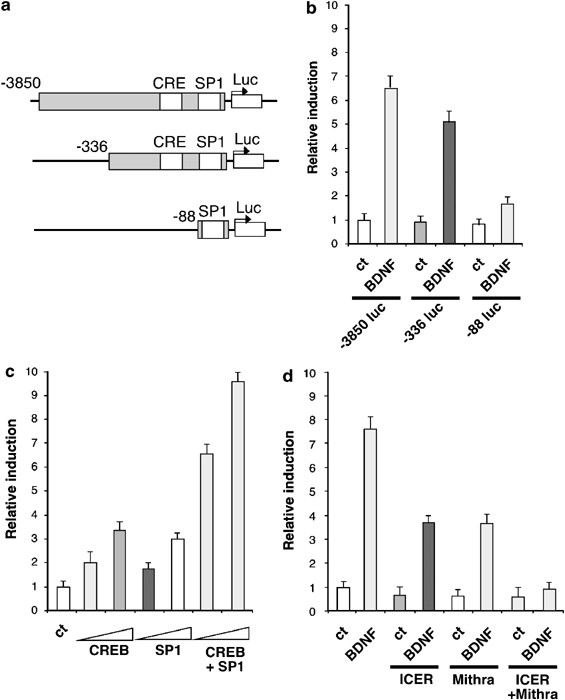
To understand how BDNF regulated the spry2 promoter, we compared a series of truncated promoters (Figure 3a). BDNF-induction remained important with the −336 promoter fragment (Figure 3b). This induction was significantly reduced with the −88 promoter fragment, but still detectable. Potential binding sites for transcription factors (AP2, ETS, NF-1, CREB, SP1) have been described in the spry2 promoter.25 Interestingly, SP1 and CREB are transcription factors regulated by the MAPK pathway.26, 27 Overexpression of either CREB or SP1 with the −336 promoter fragment stimulated its activity, and overexpression of both factors led to a synergistic induction (Figure 3c). We blocked the activity of CREB and SP1 by using respectively, ICER, a dominant negative form of CREB, and mithramycin A, a drug that inhibits the activity of the transcription factors of the SP1 family.28, 29 Separately, ICER and mithramycin reduced by approximately 50% the induction of the spry2 promoter by BDNF, whereas overexpression of ICER and concomitant treatment with mithramycin blocked BDNF-induced activity of the promoter (Figure 3d). These results suggest that BDNF stimulates spry2 expression by regulating its promoter through cooperation between CREB and SP1.
Figure 3
BDNF regulates spry2 expression through CREB and SP1. (a) Schematic representation of the spry2 promoter reporter genes containing either the region (−3850; +221 bp), (−336; +221 bp) or (−88; +221 bp). CRE and SP1 binding sites are indicated. (b) BDNF-inducibility of the truncated spry2 promoters. Neurons were transfected with the indicated luc plasmid at day 1 for 24 h, serum-deprived for 2 h and treated with BDNF (50 ng/ml) for 8 h before luc assay. (c) Neurons were co-transfected with the −336 luc spry2 reporter gene and the CREB or SP1 expression vectors as indicated. Two doses of expression vectors were used (50, 150 ng). (d) Neurons were co-transfected with the −336 luc spry2 reporter gene and the ICER expression vector or an empty vector, in presence or in absence of BDNF (50 ng/ml) and Mithramycin (25 _μ_M), an inhibitor of the SP1 family of transcription factors. In (b), (c) and (d) 24 h after transfection, cells were lysed and luc was quantified. Bars represent means±S.D. in a representative experiment performed in triplicates
Spry2 antagonizes BDNF-induced signaling pathways in neurons
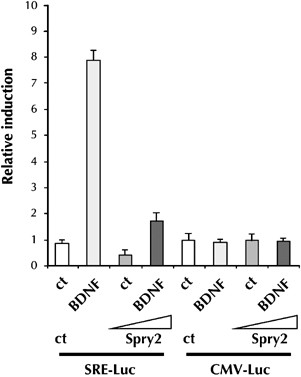
BDNF regulates gene expression in CGN and we previously showed that it leads to an increase in the activity of a serum-responsive element (SRE) in the c-fos promoter.6 Therefore, we determined whether Spry2 could inhibit BDNF-induced signaling by using a SRE-luciferase reporter gene (SRE-Luc). As previously observed, BDNF treatment stimulated the expression of the SRE-driven reporter gene (Figure 4). Importantly, overexpression of Spry2 strongly reduced this stimulation of the SRE reporter gene. A control reporter gene cytomegalovirus-Luc (CMV-Luc) was not affected by BDNF treatment or Spry2 expression. These data demonstrate that Spry2 can inhibit the transcriptional effects of BDNF and suggest that Spry2 can regulate the signaling pathways controlled by BDNF. The functional relationship between BDNF and Spry2 is also supported by the fact that BDNF treatment modifies the subcellular localization of a Spry2–YFP (Spry2–yellow fluorescent protein) fusion protein (Supplementary data S1).
Figure 4
Spry2 inhibits BDNF-induced gene expression. Immature CGN were transfected with an SRE-Luc reporter gene or a control CMV-Luc plasmid and treated with BDNF for 18 h in the presence or absence of the Spry2 expression vector. At 24 h after transfection, cells were lysed and luc was quantified. Bars represent means±S.D. in a representative experiment performed in triplicates
Overexpression of Spry2 inhibits neuritogenesis.
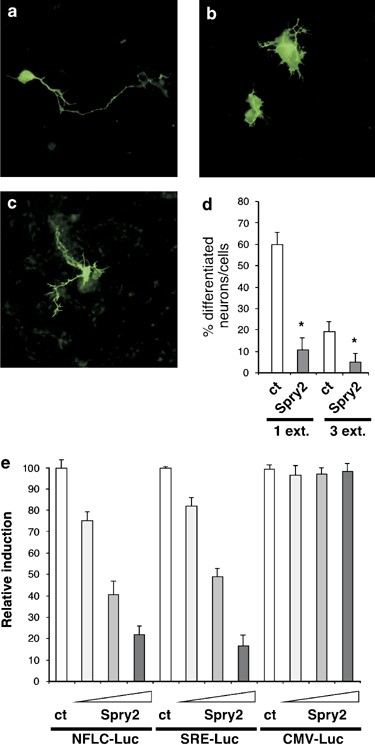
Having demonstrated that Spry2 expression can regulate BDNF-induced signaling pathways, we examined whether Spry2 overexpression during neuronal terminal maturation had an impact on neurite outgrowth. Immature neurons were co-transfected with plasmids expressing Spry2 and green fluorescent protein (GFP). After 36 h, cells were fixed and observed under a fluorescence microscope to analyze the number and the shape of the neurites of GFP-positive neurons. We identified three types of neurons depending on their neuritic extensions: (i) neurons with one or two long extensions, which predominated in the control condition (Figure 5a); (ii) neurons without extension or with short extensions (Figure 5b), which prevailed when Spry2 was overexpressed; (iii) neurons with three or more extensions with multiple ramifications (Figure 5c). Quantification confirmed that overexpression of Spry2 significantly decreased the number of neurons with one or two extensions, as well as the number of neurons with three or more ramified extensions (Figure 5d). Conversely, Spry2 increased the number of neurons with no extension. To further establish this inhibitory effect of Spry2 in maturating neurons, we examined the consequences of Spry2 expression on the transcriptional activity of the neurofilament light chain (NFLC) promoter and of the SRE-containing promoter, using luc reporter genes (NFLC-Luc and SRE-Luc, respectively). NFLC is involved in neurite outgrowth and function, and its expression correlates with morphological differentiation of neural cells.30 Transfection of increasing doses of a Spry2-expression plasmid led to a dose-dependant inhibition of both NFLC-Luc and SRE-Luc, but not of the control CMV-Luc (Figure 5e). Together, these results show that high levels of Spry2 have an inhibitory effect on neurite growth.
Figure 5
Overexpression of Spry2 inhibits neuritogenesis. (a–d) Immature CGN were co-transfected with a GFP expression vector and a Spry2 expression vector or the corresponding empty vector as control (Ct). At 36 h after transfection, neurons were fixed and observed under a fluorescence microscope. Shape and number of neurites of GFP-positive neurons were analyzed and counted. Neurons with one extended neurite were predominant following transfection of the empty expression vector (a). Neurons with short or no extension were frequently observed upon overexpression of Spry2 (b). Few neurons displayed three or more neurites (c). The graph (d) shows means±S.D. obtained in a typical experiment carried out in triplicates. Asterisks indicate significant difference (P<0.05) compared to control, as calculated by a one-way ANOVA test, followed by a Newman–Keul's test. Similar results were obtained in three independent experiments. (e) Immature CGN were co-transfected with luc reporter plasmids (NFLC-Luc, SRE-Luc or CMV-Luc; 0.25 _μ_g) and increasing amounts of a Spry2 expression vector (0.1; 0.25, 0.5 _μ_g) or the corresponding empty vector (ct). After 18 h, cells were lysed and luc activity in each condition was assessed. To facilitate comparison, luc units measured for each reporter in the absence of Spry2 were considered as 100%. Graph represents means±S.D. obtained in a typical experiment done in triplicates
Inactivation of Spry2 function favors neuritogenesis
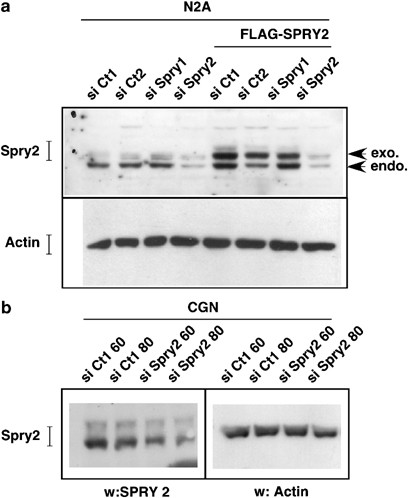
To show the involvement of endogenous Spry2 in neurite growth, we silenced its expression using RNA interference. We first verified the efficiency of the small interfering RNA (siRNA) duplex in Neuro-2a (N2A) cells co-transfected with siRNA duplexes and FLAG-tagged Spry2 expression vector. Transfection with two different control siRNAs or an siRNA specific of Spry1 did not significantly decrease the expression of exogenous or endogenous Spry2 (Figure 6a). In contrast, the Spry2 siRNA clearly reduced the amount of both exogenous and endogenous Spry2 proteins. The Spry2 siRNA also significantly reduced Spry2 expression in CGN (Figure 6b).
Figure 6
Spry2 siRNA downregulate exogenous and endogenous Spry2 in neurons. (a) N2A neuronal cells were transfected with 50 nM of a control siRNA (si Ct1, si Ct2 or si Spry1) or Spry2 siRNA (si Spry2). In the four right lanes, the Flag-Spry2 expression vector was co-transfected along the siRNA (Flag-Spry2). After 48 h, cells extracts were analyzed by immunoblotting with a Spry2 antibody (upper panel) or an Actin antibody (lower panel) to control protein loading. (b) Immature CGN were transfected with 60 or 80 nM of control siRNA (si Ct1) or Spry2 siRNA (si Spry2) for 48 h before western blot analysis with a Spry2 antibody (left panel) or an Actin antibody (right panel) to control protein loading
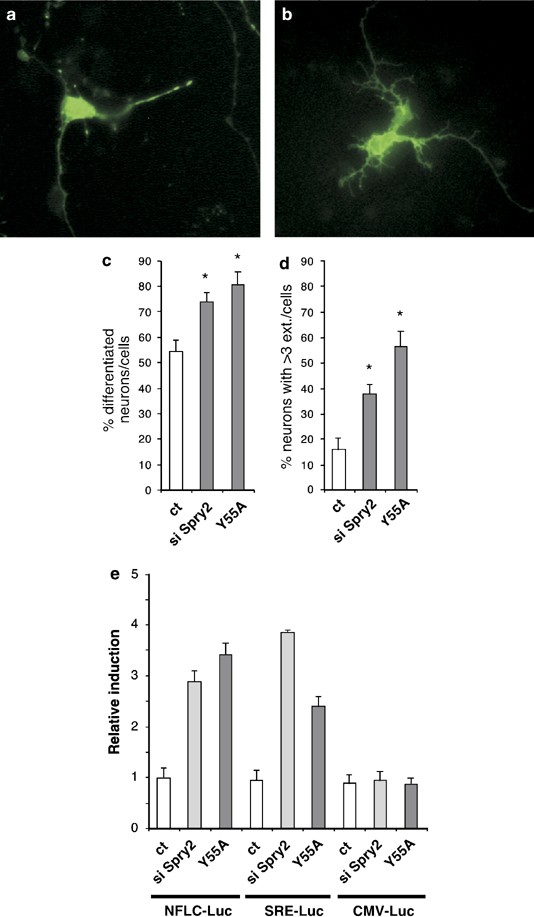
To follow the morphological changes induced by Spry2 silencing in CGN, we co-transfected the siRNA duplex along with a GFP expression vector with an estimated ratio of 4 to 1 in favor of the siRNA. After co-transfection, we examined CGN under a fluorescence microscope to assess the number and shape of the neurites. We distinguished three types of neurons: (i) neurons without extension; (ii) neurons with one or two well-developed neurites (Figure 7a); and (iii) neurons with three or more highly ramified neurites (Figure 7b). Downregulation of Spry2 expression led to an increase in the number of neurons with extensions (Figure 7c). In particular, the number of neurons with three or more neurites was significantly more common (Figure 7d). To confirm these observations, we took advantage of Spry2 Y55A, a dominant negative mutant of Spry2.24 Overexpression of Spry2 Y55A also led to a clear increase in the number of differentiated neurons (Figure 7c), especially the ones with at least three extensions (Figure 7d). Interestingly, Spry2 siRNA, as well as the Spry2 Y55A variant, increased the activity of both NFLC-Luc and SRE-Luc, but not of the control CMV-Luc reporter gene upon co-transfection (Figure 7e). Taken together, our data show that the level of expression of Spry2 in immature neurons affects signaling and gene expression involved in the control of neuritogenesis.
Figure 7
Downregulation of Spry2 favorises neuritogenesis. (a–d) Immature CGN were co-transfected for 48 h with a vector expressing the GFP and either the Spry2 siRNA duplexes or a vector expressing the Spry2 Y55A dominant-negative mutant. Pictures (a, b) show representative neurons visualized by fluorescence microscopy. GFP-positive neurons without or with (differentiated neurons) neurites were counted. Graph (c) represents means±S.D. of differentiated neurons (in %) obtained in a typical experiment done in triplicates. Graph (d) corresponds to mean±S.D. of GFP-positive neurons exhibiting three or more neurites in a typical experiment carried out in triplicates. For (c) and (d), asterisks indicate significant difference (P<0.05) compared to control, as calculated by a one-way ANOVA test followed by a Newman–Keul's test. (e) Immature CGN were co-transfected with luc reporter plasmids (NFLC-Luc, SRE-Luc or CMV-Luc; 0.25 _μ_g) and Spry2 siRNA (si Spry2, 50 nM) or a vector expressing the Spry2 Y55A dominant-negative mutant (Y55A, 0.5 _μ_g). After 18 h, cells were lysed and luc activities were assessed. To facilitate comparison, luc units measured for each reporter in the absence of Spry2 siRNA or Spry2Y55A were set at 1. Bars represent means±S.D. obtained in a typical experiment carried out in triplicates
Neurotoxic stresses downregulate Spry2 expression.
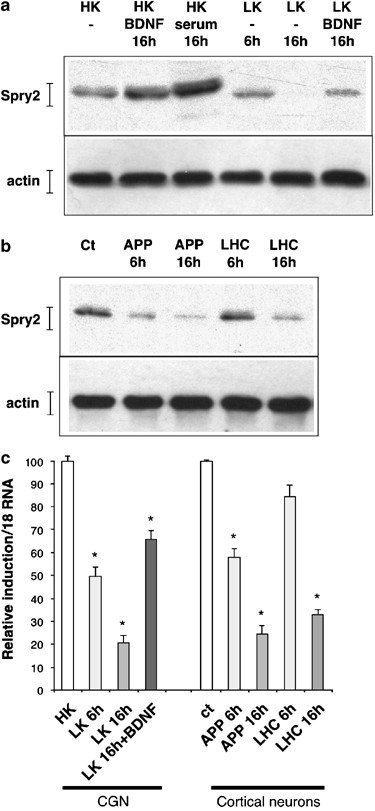
Serum and BDNF have also been involved in the survival of fully differentiated neurons.31 Therefore, we hypothesized that Spry2 might contribute to neuronal survival through the regulation of the Ras/Erk MAPK pathway or alternate pathways induced by BDNF or serum in mature neurons. We fist examined whether BDNF regulated the expression of Spry2 in differentiated CGN at day 5 of the culture, when neurons are grown in DMEM without serum. As observed in immature neurons, BDNF or serum increased Spry2 protein levels (Figure 8a). Neuronal apoptosis has been extensively studied in CGN,17 which are healthy in a medium containing 25 mM potassium (HK, high potassium) and die by apoptosis when this concentration is decreased to 5 mM (LK, low potassium). Interestingly, we observed a drastic decrease in Spry2 protein level in this apoptotic condition (LK, Figure 8a). Spry2 protein levels were partially restored by BDNF treatment (Figure 8a).
Figure 8
Neurotoxic stresses downregulate spry2 expression. (a) Mature CGN were grown in the indicated culture conditions (ser, serum: 10%; HK: 25 mM; LK: 5 mM; BDNF: 25 ng/ml) for 6 or 16 h before lysis and immunoblotting with the indicated antibodies. (b) Mature cortical neurons were submitted for 6 or 16 h to the indicated pro-apoptotic stress (APP, APP-directed antibody: 100 _μ_g/ml; LHC: 200 _μ_M). Neurons extracts were analyzed as in (a). (c) Mature CGN or mature cortical neurons were grown in the indicated culture conditions (as in a or b) before lysis. Total RNA was submitted to reverse transcription followed by real-time PCR amplification of spry2 and of 18s cDNA fragments. The graph shows spry2 relative expression after normalization with 18s levels and with spry expression in the presence of only serum set at 100. Bars represent means±S.D. in a representative experiment performed in triplicates
To extend this observation, we used primary cultures of cortical neurons and submitted them to two neurotoxic treatments: (i) an antibody directed against the extracellular fragment of the APP, which induces the APP-signaling pathway and neuronal cell death;32 (ii) L-homo-cystein (LHC).33 In these neurotoxic conditions, the protein levels of Spry2 were significantly decreased (Figure 8b). Analysis of the mRNA levels by RT-qPCR showed that all the stresses that induced neuronal cell death led also to a marked decrease of spry2 mRNA (Figure 8c). Thus, neurotoxic stresses appear to lead to a reduction of spry2 expression in mature neurons.
Spry2 contributes to neuronal apoptosis
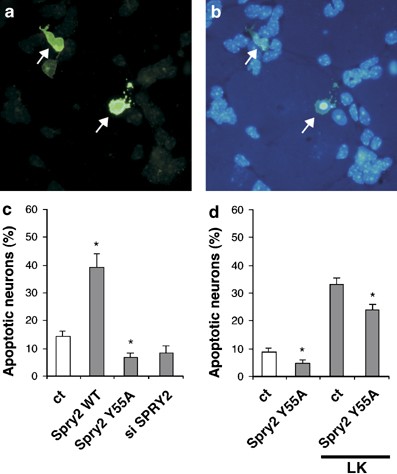
As expression of Spry2 is downregulated during neuronal cell death, we investigated whether Spry2 was involved in neuronal survival. We modified Spry2 activity in mature neurons by overexpressing wild-type or Y55A dominant-negative mutant Spry2, or by silencing its expression with Spry2 siRNA. Cells were also co-transfected with the GFP expression vector and nuclei were stained with Hoechst 3233. GFP-positive neurons were identified by fluorescence microscopy (Figure 9a) and the shape of their nuclei was examined. Condensed or fragmented nuclei were counted as apoptotic nuclei (Figure 9b). In the control condition, between 5 and 15% of neurons underwent apoptosis (Figure 9c). Overexpression of wild-type Spry2 increased more than twofold the number of apoptotic neurons. Inversely, inhibition of Spry2 activity with the Y55A dominant-negative mutant or silencing Spry2 expression with the siRNA led to a reduction of apoptotic neurons (Figure 9c). Downregulation of Spry2 activity also caused a decrease of neuronal apoptosis in LK condition (Figure 9d). Thus, the activity of Spry2 impacts on the survival of mature neurons.
Figure 9
Spry2 contributes to neuronal apoptosis. Mature CGN were co-transfected for 48 h with an expression vector for GFP and an expression vector for wild-type Spry2 (WT) or Spry2 Y55A (Y55A) or the Spry2 siRNA (si Spry2). After fixation and staining with Hoechst 3343 dye, GFP-positive cells (a, arrows) were identified by fluorescence microscopy and the morphology (intact, fragmented or condensed) of their nucleus was examined (b, arrows). (c, d) Percentages of apoptotic neurons among the GFP-positive cells were quantified and presented as means±S.D. obtained in a typical experiment done in triplicate. Asterisks indicate significant difference (P<0.05) compared to control, as calculated by a one-way ANOVA test followed by a Newman–Keul's test. (d) At 24 h after transfection, cells were switched to LK medium for 24 h before fixation and staining
Discussion
Neuronal differentiation and survival are involved in the development and the proper activity of the CNS. Neurotrophins play key roles in the regulation of these processes by binding to RTK and activating signaling cascades such as the Ras/Erk MAPK. Here, we investigated for the first time the regulation and the function of Spry2, the prototypical member of a new family of RTK signaling modulators, in primary cultures of neurons from the CNS. We found that the expression of Spry2 is progressively increased during in vitro differentiation of CGN, is positively regulated by the neurotrophin BDNF and negatively regulated by various neurotoxic stresses. We also demonstrated that Spry2 expression affects the neuronal physiology by establishing an inhibitory-feedback loop of BDNF activity, reducing neuritogenesis and survival.
Spry2 belongs to a negative-feedback loop that controls BNDF activity in neurons
In the mouse embryo, Spry2, as well as Spry1 but not Spry4, are expressed in a transverse domain that encompasses the prospective caudal midbrain, isthmus and cerebellum around embryonic days (E) 9.5–10.5.15 We found that Spry2, as well as Spry1 (data not shown), were also expressed in the brain of 5 days old mice, especially in the cerebellum (Figure 1c), the hippocampus (Figure 1b) and the cortex (Figure 1a). In primary cultures, expression of Spry2 in differentiated CGN was confirmed by its colocalization with the neuronal marker MAP2 (Figure 1i, j). Remarkably, Spry2 expression was increased in parallel to neuronal differentiation of CGN (Figures 1d, 10). In cancer cell lines, the amount of Spry is regulated by serum or growth factors such as FGF.22 In this study, we found that the neurotrophin BDNF stimulated Spry2 expression in neurons (Figures 2a, 10). Positive regulation of Spry2 by BDNF treatment or during neuronal differentiation may appear intriguing, as we also showed that Spry2 antagonizes BDNF-induced signaling (Figures 4, 10) and neuronal differentiation (Figures 5, 10). However, by regulating the level of their own antagonist in a negative-feedback loop, growth factors or neurotrophins become able to limit precisely the strength, duration and range of signaling, which is critical for the induction of the correct physiological outcome.
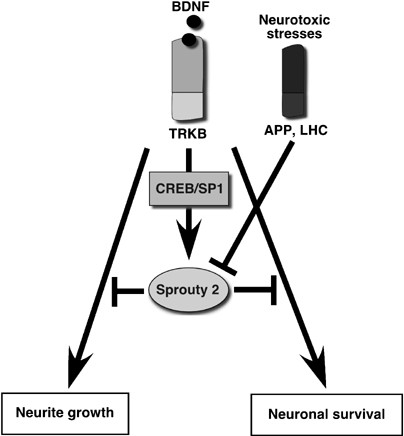
Figure 10
Involvement of Spry2 in neuronal survival and differentiation. Expression of Spry2 is regulated by BDNF in CGN during the terminal differentiation and when the neuron is differentiated. Spry2 inhibits BDNF-induced transduction mechanisms and participates to the control of neurite growth and neuronal survival. Therefore Spry2 is involved in a negative-feedback loop initiated by BDNF and that control BDNF effect on neurite growth and neuronal survival
Further investigation of the mechanism involved revealed that transcriptional activity of the spry2 promoter was increased by BDNF (Figure 2c, d). A direct stimulation of two pathways induced by BDNF, the Erk1 MAPK and the Akt pathway, also activated the spry2 promoter (Figure 2d). Similarly, Osaki reported that growth factor-induced activation of spry1 transcription was dependent on the Erk1/2 pathway.23 Finally, we identified CREB and SP1 as two transcription factors that cooperate to regulate spry2 promoter activity in neurons in response to BDNF (Figures 3, 10).
Spry participates to the control of neurite growth
Spry proteins have been implicated in many developmental processes, but so far only a few studies examined the role of Spry2 in neural cells. Loss of spry2 in mice resulted in enteric nerve hyperplasia due to excessive GDNF signaling.34 In the chick embryo, Spry2 expression in the isthmus was shown to contribute to the regionalization of the presumptive mesencephalon and metencephalon, the latter giving rise to the future cerebellum.35 The data obtained in this study suggest that Spry2 also participates to the control of neurite growth.
Depending on the type of neurons (granules, pyramidal, motoneurons and so on), neurites will vary in their length, number, ramification and number of spikes. The extracellular signals and their intracellular relays that control such orchestrated complexity are still largely enigmatic. Growth factors and neurotrophins participate to these events.1 The Ras/Erk MAPK pathway and other pathways controlled by Cdc42 and Rac GTPases also modulate neurite growth.36 However, how these signals work together in a given cell to determine when and where the neurite, or a ramification, has to grow, and then, when and where, it has to stop, is not yet well understood. Here, we propose that Spry2 expression is involved in the control of neuritic growth (Figure 10). First, Spry2 expression increases progressively until neurons are differentiated, correlating with neurite growth (Figure 1d). Second, modulation of Spry2 expression interferes with neurite growth: overexpression of Spry2 inhibits neurite growth (Figure 5), whereas inhibition of Spry2 activity by RNA interference or expression of a dominant-negative mutant favors neuritogenesis (Figure 7). These effects were also observed on the neurite growth induced by Rac or Cdc42 (Supplementary data S2). These results are in agreement with a previous study where ectopic expression of Spry proteins in pheochromocytoma PC12 cells blocked neurite growth and branching induced by NGF or bFGF.22 In the embryonic kidney or the lung, Spry proteins play key roles during branching morphogenesis processes.12 Likewise, expression of Spry in a feedback loop may allow the right dose of signaling required for appropriate neurite growth and branching.
The exact molecular mechanism of Spry2 activity has not yet been clarified. Depending on the cell type and/or the growth factor studied, Spry proteins were shown to interfere with the Ras/Erk MAPK signaling pathway at the level of Ras or Raf.8, 9, 22 Spry2 seems to localize at the membrane and in the neurites (Supplementary data S1), which is consistent with an inhibition of early steps of signaling, at the level of small GTPases or Raf.
Role of Spry2 in neuronal survival
In differentiated neurons, we showed that overexpression of Spry2 favored neuronal apoptosis, whereas its downregulation had a neuroprotective effect, even under neurotoxic stresses (Figures 9, 10). This observation is consistent with the increase in the number of neurons caused by the loss of spry in Drosophila.37 In contrast, in immature CGN, Spry2 expression controls neuritogenesis and seems to have little effect on neuronal survival (Figures 5, 7). Reciprocally, we did not detect any significant differences in the neurite network, when Spry2 was modified in mature neurons (data not shown). The causes of the differences observed between immature and mature neurons are not cleat yet.
The fact that Spry2 antagonizes neuronal survival, whereas being increased during neuronal differentiation is surprising. Note that Spry proteins have yet not been linked directly to any effectors of apoptosis. The apoptosis observed here might rather be due to the inhibition of a pathway promoting neuronal survival such as the PI3-K/Akt pathway.38 Therefore, it is possible that the amount of Spry2 present in the neurons allows fine-tuning of the transduction pathways controlling neuronal survival. Indeed, the sustained expression of Spry2 in mature neurons might be critical to allow the equilibrium between death and survival. This equilibrium permits the correct development of a neuronal network in which only the appropriately connected neurons survive. Inhibition of Spry2 by siRNA or use of a dominant-negative mutant may shift the equilibrium towards survival by increasing the activity of pro-survival pathways, whereas production of high amounts of Spry2 by transient transfection would shift the equilibrium towards death.
In this context, the loss of Spry expression induced by neurotoxic stress is also surprising given that Spry2 overexpression favors apoptosis (Figures 9, 10). Certainly, the loss of Spry2 is not the mechanism that mediates neuronal cell death induced by the neurotoxic stresses. On the contrary, the loss of the negative-feedback loop of the neurotrophic signals by a yet unknown mechanism might represent a compensatory process that would enhance the ability of the neuron to survive.
Materials and Methods
Cell culture
Primary cultures of CGN were obtained from 7 days old mice as described previously.6 Cortical neurons were dissected from E16 murine embryos. Briefly, after enzymatic and mechanical dissociation, cells were plated at a density of 500 cells/mm2 on 0.1 mg/ml polyornithine-pre-coated culture dishes and were grown at 37°C in a humidified atmosphere (5% CO2/95% air). For CGN, plating culture medium contained DMEM (Life Technology) supplemented with 10% heat-inactivated horse serum (Life Technology), 100 nM insulin (Life Technology), 50 mg/ml gentamycin (Life Technology) and 25 mM KCl. At 48 h after plating, cells were switched to defined medium (called HK medium) containing DMEM supplemented with 10 nM insulin, 100 mg/ml human transferrin, 60 mM putrescine, 30 nM sodium selenite, 50 mg/ml gentamycin and 25 mM KCl (final [K+]=30 mM). LK medium was obtained by omitting KCl (final [K+]=5 mM). For cortical neurons, culture media were identical except for KCl (final [K+]=5 mM). Neurons were considered as immature up to 2 days after plating and as mature 5 days after plating. Murine N2A cells were obtained from ATCC and maintained in DMEM supplemented with 10% fetal bovine serum and antibiotics.
Vectors, siRNA and reagents
Mammalian expression vectors for Flag-tagged Spry2 (wild type or Y55A) were described previously.22, 24 The YFP–Spry2 and GFP–Spry2 vectors were constructed by fusion of the GFP or YFP moieties to the N terminus of the open reading frame. The SRE-Luc reporter plasmid was a gift from Dr. R. Prywes (Columbia University, NY, USA) and contains five repeats of the SRE upstream of a c-fos minimal promoter and the Luc reporter gene. The NFLC-Luc reporter plasmid contains the promoter fragment of the NFLC (−650; +82) fused to the Luc reporter gene and was from Dr. L.E. Heasley (University of Colorado Health Sciences Center, CO, USA).39 CMV-Luc contains a minimal CMV promoter upstream the luc reporter gene. Mammalian expression vectors for Rac1 and Cdc42 were from Dr. L. Van Aelst (Cold Spring Harbor Laboratory, NY, USA). The spry2 promoter constructs25 were from Dr. D. Warburton (Childrens Hospital Los Angeles Research Institute, CA, USA): full-length −3850 luc (−3850; +221); −336 luc (−336; +221); −88 luc (−88; +221). The CREB expression vector was previously described.6
Stealth siRNA annealed duplexes were obtained from Invitrogen and targeted the following 5′ to 3′ DNA sequences: si Spry2, GCCATCCGAAACACCAATGAGTACA; si Spry1, GCAGGAAAGGACTCATGAAATCATA; si Ct1, sequence control GC 50% (Invitrogen); si Ct2, GCAGAGCATGTGACCGACATTGTTA.
BDNF, LHC and Mithramycin were purchased from Sigma.
Transfections
Immature neurons were dissected and allowed to attach to culture plates for 4 h before transfection. Mature neurons were cultured for 4–5 days before transfection. Gene transfer was performed using polyethyleneimine (PEI) as a DNA carrier.6 Neurons were transfected with 0.33 _μ_l PEI/_μ_g of DNA and centrifuged for 5 min at 1500 r.p.m. (400 g). After 30 min, neurons were transferred to fresh medium and assays were performed 20–48 h later. For GFP co-transfection, 0.2 _μ_g of GFP-encoding vector was added to the other vectors. For luc assays, cells were lysed 24 h after transfection using the luc assay lysis reagent (Promega) and cell extracts were mixed with D-luciferin (Sigma). Luc activity was measured by a luminometer (Berthold systems). For siRNA transfection, Lipofectamine 2000 (Invitrogen) was used as recommended, but the culture plate was centrifuged for 5 min at 1500 r.p.m. after addition of the transfection mix.
Immunocytochemistry, Hoechst staining and fluorescence microscopy
Cells were fixed with 4% paraformaldehyde (PAF) in phosphate-buffered saline (PBS) for 30 min at room temperature. Cells were then permebilized and saturated with PBS containing 1% NP40 and 3% horse serum. Primary antibodies (Spry2 and MAP2) were added in the same solution at a concentration of 1/500 for 24 h. After three PBS washes, Alexa secondary antibodies (Molecular Probes; 1/2000) were added for 2 h. Cells were then washed three times and incubated for 30 min with the nuclear Hoechst 3343 dye (Sigma 1 _μ_g/ml). Fixed cells were viewed (Hoechst staining, 420 nm; GFP fluorescence, Spry2 staining, 510 nm; MAP2 staining, 575 nm) on a Nikon microscope (E800) equipped with a Nikon digital camera (DXM1200) linked to image acquisition software (Lucia, Nikon). Confocal microscopy was performed on a Nikon microscope (Diaphot 300) equipped by a Bio-Rad confocal system (MRC1024).
Immunoblot
Cells cultured in six-well plates were lysed in 120 _μ_l of Laemmli's buffer (125 mM Tris/HCl (pH 6.7), containing 3.3% sodium dodecyl sulfate (SDS), 0.7 M 2-mercaptoethanol, 10% glycerol and 0.02% Bromophenol Blue). Extracts were sonicated for 20 s and boiled for 5 min before centrifugation at 20 000 × g for 5 min. Proteins (∼60 _μ_g) were separated on 10% SDS-polyacrylamide gel electrophoresis, electrotransferred to nitrocellulose membranes which were blocked with fat-free milk (5% in PBS) and incubated with the various primary antibodies (mouse anti-Actin 1/200, gift from Dr. Aunis, INSERM U.575, France; rabbit anti-Spry2 1/2000; mouse anti-MAP2 clone MT−01 1/1000, Santa Cruz; mouse anti-APP Mab348 1/1000, Chemicon) and a relevant secondary horseradish-peroxidase-conjugated antibody (Sigma, 1/2000). Finally, membranes were developed by enhanced chemiluminescence detection.
Quantitative RT-PCR
Cells were grown on 10 cm dishes. After treatments, they were harvested and total RNA was extracted with the Qiagen RNeasy kit. Reverse transcription was performed with 1 _μ_g RNA using iScript cDNA Synthesis kit (Bio-Rad). Real-time-qPCR was performed with an iCycler (Bio-Rad) using SYBR Green dye (iQ SYBR green Supermix, Bio-Rad). For each gene, a standard curve based on successive cDNA dilutions was performed and used to calculate starting quantities of the subsequent cDNA. To ensure a thorough calculation, starting quantities of cDNAs of interest were compared to those of a house-keeping gene (18s) in the same plate. After each qPCR, specificity of the amplification was controlled by a melting curve ranging from 55–95°C, whereby a single peak corresponding to the amplicon was present.
All primers were designed in regions flanking introns to exclude data alteration by possible DNA contamination. Spry2 forward primer: 5′-GGC TTT CAG AAC TAT GAG CCA AAT-3′; spry2 reverse primer: 5′-ATT TGG CTC ATA GTT CTG AAA GCC-3′; 18s forward primer: 5′-CGT CTG CCC TAT CAA CTT TCG-3′; 18s reverse primer: 5′-TTC CTT GGA TGT GGT AGC CG-3′.
RNA in situ hybridization
Brains of 5 days old mouse pups were dissected and fixed in PAF 4% overnight at 4°C. After incubation in 30% Sucrose overnight at 4°C, brains were embedded and kept at −80°C until sectioning. RNA ISH was carried out on 12 _μ_m cryostat sections as described previously.40 Briefly digoxigenin (dig)-labeled RNA probes were generated using T3 or T7 RNA polymerases and a dig-RNA-labelling mix (Roche). Probes were hybridized to the slides overnight at 65°C in a buffer composed of 50% formamide, 200 mM sodium, chloride, 20 mM sodium acetate, 10% dextran sulfate and 1 mg/ml tRNA, then washed twice at 65°C in 50% formamide, 150 mM sodium chloride, 15 mM sodium citrate, 0.1% Tween 20 and washed two times in MABT buffer. Sections were next incubated with an alkaline phosphatase-coupled anti-dig antibody (Roche), washed 5 times in MABT buffer. Specific sense and antisense dig probes were obtained from Spry plasmids as described previously.11
Abbreviations
APP:
amyloid beta precursor protein
BDNF:
brain derived neurotrophic factor
CGN:
cerebellar granule neurons
CMV:
cytomegalovirus
CNS:
central nervous system
dig:
digoxigenin
DMEM:
Dulbecco's modified Eagle's medium
E:
embryonic day
EGL:
external granule cell layer
FGF:
fibroblast growth factor
GDNF:
glial cell derived neurotrophic factor
GFP:
green fluorescent protein
HK:
high potassium
ISH:
in situ hybridization
LHC:
L-homo-cystein
LK:
low potassium
Luc:
luciferase
MAPK:
mitogen-activated protein kinase
NFLC:
neurofilament light chain
PAF:
paraformaldehyde
PBS:
phosphate-buffered saline
PEI:
polyethyleneimine
RTK:
receptor tyrosine kinase
RT-qPCR:
reverse transcription-quantitative polymerase chain reaction
siRNA:
small interfering RNA
YFP:
yellow fluorescent protein
References
- Lewin GR, Barde YA . Physiology of the neurotrophins. Annu Rev Neurosci 1996; 19: 289–317.
Article CAS Google Scholar - Tanaka S, Sekino Y, Shirao T . The effects of neurotrophin−3 and brain-derived neurotrophic factor on cerebellar granule cell movement and neurite extension in vitro. Neuroscience 2000; 97: 727–734.
Article CAS Google Scholar - Poo MM . Neurotrophins as synaptic modulators. Nat Rev Neurosci 2001; 2: 24–32.
Article CAS Google Scholar - Thoenen H, Barde YA, Davies AM, Johnson JE . Neurotrophic factors and neuronal death. Ciba Found Symp 1987; 126: 82–95.
CAS PubMed Google Scholar - Arevalo JC, Wu SH . Neurotrophin signaling: many exciting surprises!. Cell Mol Life Sci 2006; 63: 1523–1537.
Article CAS Google Scholar - Gaiddon C, Loeffler JP, Larmet Y . Brain-derived neurotrophic factor stimulates AP-1 and cyclic AMP-responsive element dependent transcriptional activity in central nervous system neurons. J Neurochem 1996; 66: 2279–2286.
Article CAS Google Scholar - Iwanami M, Hiromi Y, Okabe M . Cell-type specific utilization of multiple negative feedback loops generates developmental constancy. Genes Cells 2005; 10: 743–752.
Article CAS Google Scholar - Mason JM, Morrison DJ, Basson MA, Licht JD . Sprouty proteins: multifaceted negative-feedback regulators of receptor tyrosine kinase signaling. Trends Cell Biol 2006; 16: 45–54.
Article CAS Google Scholar - Kim HJ, Bar-Sagi D . Modulation of signalling by Sprouty: a developing story. Nat Rev Mol Cell Biol 2004; 5: 441–450.
Article CAS Google Scholar - Mailleux AA, Tefft D, Ndiaye D, Itoh N, Thiery JP, Warburton D et al. Evidence that SPROUTY2 functions as an inhibitor of mouse embryonic lung growth and morphogenesis. Mech Dev 2001; 102: 81–94.
Article CAS Google Scholar - Gross I, Morrison DJ, Hyink DP, Georgas K, English MA, Mericskay M et al. The receptor tyrosine kinase regulator Sprouty1 is a target of the tumor suppressor WT1 and important for kidney development. J Biol Chem 2003; 278: 41420–41430.
Article CAS Google Scholar - Basson MA, Akbulut S, Watson-Johnson J, Simon R, Carroll TJ, Shakya R et al. Sprouty1 is a critical regulator of GDNF/RET-mediated kidney induction. Dev Cell 2005; 8: 229–239.
Article CAS Google Scholar - Klein OD, Minowada G, Peterkova R, Kangas A, Yu BD, Lesot H et al. Sprouty genes control diastema tooth development via bidirectional antagonism of epithelial-mesenchymal FGF signaling. Dev Cell 2006; 11: 181–190.
Article CAS Google Scholar - Chi L, Itaranta P, Zhang S, Vainio S . Sprouty2 is involved in male sex organogenesis by controlling fibroblast growth factor 9-induced mesonephric cell migration to the developing testis. Endocrinology 2006; 147: 3777–3788.
Article CAS Google Scholar - Minowada G, Jarvis LA, Chi CL, Neubuser A, Sun X, Hacohen N et al. Vertebrate Sprouty genes are induced by FGF signaling and can cause chondrodysplasia when overexpressed. Development 1999; 126: 4465–4475.
CAS PubMed Google Scholar - Zhang S, Lin Y, Itaranta P, Yagi A, Vainio S . Expression of Sprouty genes 1, 2 and 4 during mouse organogenesis. Mech Dev 2001; 109: 367–370.
Article CAS Google Scholar - Contestabile A . Cerebellar granule cells as a model to study mechanisms of neuronal apoptosis or survival in vivo and in vitro. Cerebellum 2002; 1: 41–55.
Article CAS Google Scholar - Izant JG, McIntosh JR . Microtubule-associated proteins: a monoclonal antibody to MAP2 binds to differentiated neurons. Proc Natl Acad Sci USA 1980; 77: 4741–4745.
Article CAS Google Scholar - Dichgans M, Monning U, Konig G, Sandbrink R, Masters CL, Beyreuther K . APP expression in primary neuronal cell cultures from P6 mice during in vitro differentiation. Dementia (Basel, Switzerland) 1993; 4: 301–307.
CAS Google Scholar - Hertz L, Schousboe I, Hertz L, Schousboe A . Receptor expression in primary cultures of neurons or astrocytes. Progress in neuro-Psychopharmacology & Biological Psychiatry 1984; 8: 521–527.
Article CAS Google Scholar - Williams EJ, Furness J, Walsh FS, Doherty P . Characterisation of the second messenger pathway underlying neurite outgrowth stimulated by FGF. Development 1994; 120: 1685–1693.
CAS PubMed Google Scholar - Gross I, Bassit B, Benezra M, Licht JD . Mammalian sprouty proteins inhibit cell growth and differentiation by preventing ras activation. J Biol Chem 2001; 276: 46460–46468.
Article CAS Google Scholar - Ozaki K, Kadomoto R, Asato K, Tanimura S, Itoh N, Kohno M . ERK pathway positively regulates the expression of Sprouty genes. Biochem Biophys Res Commun 2001; 285: 1084–1088.
Article CAS Google Scholar - Mason JM, Morrison DJ, Bassit B, Dimri M, Band H, Licht JD et al. Tyrosine phosphorylation of Sprouty proteins regulates their ability to inhibit growth factor signaling: a dual feedback loop. Mol Biol Cell 2004; 15: 2176–2188.
Article CAS Google Scholar - Ding W, Bellusci S, Shi W, Warburton D . Functional analysis of the human Sprouty2 gene promoter. Gene 2003; 322: 175–185.
Article CAS Google Scholar - Xing J, Kornhauser JM, Xia Z, Thiele EA, Greenberg ME . Nerve growth factor activates extracellular signal-regulated kinase and p38 mitogen-activated protein kinase pathways to stimulate CREB serine 133 phosphorylation. Mol Cell Biol 1998; 18: 1946–1955.
Article CAS Google Scholar - Merchant JL, Du M, Todisco A . Sp1 phosphorylation by Erk 2 stimulates DNA binding. Biochem Biophys Res Commun 1999; 254: 454–461.
Article CAS Google Scholar - Ryu H, Lee J, Hagerty SW, Soh BY, McAlpin SE, Cormier KA et al. ESET/SETDB1 gene expression and histone H3 (K9) trimethylation in Huntington's disease. Proc Natl Acad Sci USA 2006; 103: 19176–19181.
Article CAS Google Scholar - Ray R, Snyder RC, Thomas S, Koller CA, Miller DM . Mithramycin blocks protein binding and function of the SV40 early promoter. The Journal of Clinical Investigation 1989; 83: 2003–2007.
Article CAS Google Scholar - Julien JP . Neurofilament functions in health and disease. Curr Opin Neurobiol 1999; 9: 554–560.
Article CAS Google Scholar - Lindholm D, Dechant G, Heisenberg CP, Thoenen H . Brain-derived neurotrophic factor is a survival factor for cultured rat cerebellar granule neurons and protects them against glutamate-induced neurotoxicity. Eur J Neurosci 1993; 5: 1455–1464.
Article CAS Google Scholar - Mbebi C, See V, Mercken L, Pradier L, Muller U, Loeffler JP . Amyloid precursor protein family-induced neuronal death is mediated by impairment of the neuroprotective calcium/calmodulin protein kinase IV-dependent signaling pathway. J Biol Chem 2002; 277: 20979–20990.
Article CAS Google Scholar - Langmeier M, Folbergrova J, Haugvicova R, Pokorny J, Mares P . Neuronal cell death in hippocampus induced by homocysteic acid in immature rats. Epilepsia 2003; 44: 299–304.
Article CAS Google Scholar - Chi L, Zhang S, Lin Y, Prunskaite-Hyyrylainen R, Vuolteenaho R, Itaranta P et al. Sprouty proteins regulate ureteric branching by coordinating reciprocal epithelial Wnt11, mesenchymal Gdnf and stromal Fgf7 signalling during kidney development. Development 2004; 131: 3345–3356.
Article CAS Google Scholar - Suzuki-Hirano A, Sato T, Nakamura H . Regulation of isthmic Fgf8 signal by sprouty2. Development 2005; 132: 257–265.
Article CAS Google Scholar - Nikolic M . The role of Rho GTPases and associated kinases in regulating neurite outgrowth. Int J Biochem Cell Biol 2002; 34: 731–745.
Article CAS Google Scholar - Kramer S, Okabe M, Hacohen N, Krasnow MA, Hiromi Y . Sprouty: a common antagonist of FGF and EGF signaling pathways in Drosophila. Development 1999; 126: 2515–2525.
CAS PubMed Google Scholar - Dudek H, Datta SR, Franke TF, Birnbaum MJ, Yao R, Cooper GM et al. Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science 1997; 275: 661–665.
Article CAS Google Scholar - Zentrich E, Han SY, Pessoa-Brandao L, Butterfield L, Heasley LE . Collaboration of JNKs and ERKs in nerve growth factor regulation of the neurofilament light chain promoter in PC12 cells. J Biol Chem 2002; 277: 4110–4118.
Article CAS Google Scholar - Myat A, Henrique D, Ish-Horowicz D, Lewis J . A chick homologue of Serrate and its relationship with Notch and Delta homologues during central neurogenesis. Dev Biol 1996; 174: 233–247.
Article CAS Google Scholar
Acknowledgements
We thank Dr. R. Prywes, Dr. L.E. Heasley and Dr. D. Warburton for the generous gift of plasmids. We thank Marie-Jose Ruivo for her excellent technical assistance. This work was supported by CNRS, ARC (no 3288), La Ligue contre le Cancer (Comité du Bas-Rhin), ANR, INSERM. I.G. is a fellow of the Institut National du Cancer (Cancéropôle Grand-Est). S.B. is a fellow of INSERM and Région Alsace.
Author information
Authors and Affiliations
- U682 INSERM, Strasbourg, France
I Gross, J-N Freund & M Kedinger - Institute of Toxicology and Genetics, Forschungszentrum, D-76021, Karlsruhe, Germany
O Armant - U692 INSERM, Université Louis Pasteur, Faculté de Médecine, Strasbourg, France
S Benosman, J L G de Aguilar, C Gaiddon & J-P Loeffler - Northwestern University Feinberg School of Medicine, Chicago, IL, USA
J D Licht
Authors
- I Gross
You can also search for this author inPubMed Google Scholar - O Armant
You can also search for this author inPubMed Google Scholar - S Benosman
You can also search for this author inPubMed Google Scholar - J L G de Aguilar
You can also search for this author inPubMed Google Scholar - J-N Freund
You can also search for this author inPubMed Google Scholar - M Kedinger
You can also search for this author inPubMed Google Scholar - J D Licht
You can also search for this author inPubMed Google Scholar - C Gaiddon
You can also search for this author inPubMed Google Scholar - J-P Loeffler
You can also search for this author inPubMed Google Scholar
Corresponding authors
Correspondence toI Gross or C Gaiddon.
Additional information
Edited by N Bazan
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Supplementary information
Rights and permissions
About this article
Cite this article
Gross, I., Armant, O., Benosman, S. et al. Sprouty2 inhibits BDNF-induced signaling and modulates neuronal differentiation and survival.Cell Death Differ 14, 1802–1812 (2007). https://doi.org/10.1038/sj.cdd.4402188
- Received: 30 January 2007
- Revised: 26 April 2007
- Accepted: 04 May 2007
- Published: 29 June 2007
- Issue Date: October 2007
- DOI: https://doi.org/10.1038/sj.cdd.4402188