The spike protein of severe acute respiratory syndrome (SARS) is cleaved in virus infected Vero-E6 cells (original) (raw)
INTRODUCTION
The severe acute respiratory syndrome (SARS), caused by SARS-associated coronavirus (SARS-CoV) 1, was recently identified as a new clinical entity. It apparently began in Guangdong province of China in November of 2002 and has spread to several countries 2. More than 8000 probable SARS cases and 900 related deaths were reported. SARS spread has caused the unstability for the globe society and dramatically decreased the economy development over the world. Accurate diagnosis and explorirg a tool to understand the pathogenesis of the disease are in urgent need 3.
SARS-CoV is now identified as a member of the coronavirus family of enveloped, positive-stranded RNA viruses, which have a broad host range. The genome of SARS-CoV has 29,727 nucleotides in length containing 11 open reading frames. The structural proteins of SARS- CoV include: the surface spike (S) glycoprotein, the membrane (M) protein, the small envelope (E) glycoprotein and the nucleocapsid (N) protein 4, 5. All of them are crucial to form the architecture of SARS CoV, especially S and N protein, which may be the best candidates for therapeutic target of SARS.
The spike protein, forming the ∼180 kD glycoprotein projection on the viral surface, is crucial for viral attachment and entry into the host cell. The first step of the infectious cycle of enveloped viruses is the interaction with cellular receptors and subsequent fusion with the cell membrane, which is mediated by spike protein. Variations of S protein among strains of coronavirus are responsible for host range and tissue tropism 6. The spike proteins of some coronaviruses have to be cleaved to become fusion competent (S1 and S2 subunits), but some others do not 7. However there is no experimental evidence to indicate that whether the spike protein of SARS-CoV is cleaved.
Nucleocapid (N) protein has multiple functions that are involved in providing nuclear-import signals, interfering cell process, virus replication and RNA package. In addition, N protein in many coronaviruses is highly conserved, immunogenic, and abundantly expressed during infection. These features make it to be a suitable candidate for raising neutralizing antibodies and diagnosis applications 8, 9.
E protein is associated with coronavirus envelope formation. Coexpression of coronavirus M protein and E protein results in the production of virus-like particles (VLPs), while expression of M protein alone does not produce VLPs 10, 11. Another major biological function of E protein is that it can induce apoptosis in E-protein-expressing cells 12. Coronavirus replicase gene encompasses two overlapping translation products, polyprotein 1a (∼450 kD) and 1b (∼750 kD), both of which are conserved in length and amino acid sequence with other coronavirus replicase proteins. Polyprotein 1a and 1b are cleaved by the internally encoded 3C-like protease (3CL) to release functional proteins necessary for virus replication 13. The SARS 3CL is fully conserved among all released SARS coronavirus genome sequences and is highly homologous with other coronavirus 3CL protease.
In order to study the functions of proteins of SARS coronavirus, specific antibodies against these proteins are in great requisites. Now we generated specific antibodies against nucleocapsid (N), spike (S1 and S2), small envelope (E) proteins and 3CL protease. Our results showed that all of these antibodies had high affinity to bind to their relevant prokaryotic expressed proteins. Furthermore, antibodies against S1, S2, N protein could detect their corresponding proteins from the lysates of SARS-CoV infected Vero E6 cells. Immunnoprecipitation showed that S protein is cleaved into S1 and S2 fragments in SARS-CoV infected Vero E6 cells, thus demonstrating the cleavage of S protein in host cells, which may play a crucial role in pathogenesis of SARS infection.
MATERIALS AND METHODS
Animals
New Zealand rabbits were purchased form Shanghai Laboratory Animal Center, Chinese Academy of Sciences. Animals were kept in conventional conditions and were handled in compliance with Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China) guidelines for animal care and use.
Prokaryotic expression and purification of S1, S2, N, E, 3-CL proteins of SARS-CoV
SARS-S1 (1-667 aa), S2 (668-1255 aa), S, E genes were in-framely inserted into vectors pGEX-4T1, pGEX-3x, pGEX-4T1 and pGEX-2T, and other two genes (N and 3CL genes) were inserted into vectors pets and then transformed into E. coli strain, BL21 (DE3). The bacterial cells were grown in LB medium at 37°C with ampicillin (1.0 mM). For the expression of SARS-S1, S2, S proteins, bacterial cells were induced by IPTG (final concentration 0.1 mM-0.2 mM) for 3 h in 22°C and then cells were recovered and disrupted by French Pressure cell press. Since most of S, S1 and S2 proteins were found in the insoluble fraction of the cell lysates, we purified S, S1, S2 proteins by Model 422 Electro-Eluter (Bio-Rad) according to Manufacturer's manual. For the expression of SARS-E protein, bacterial cells were induced with 0.5 mM of IPTG. After induction at 25°C for 5 h, cells were harvested by centrifugation and suspended in 20 ml of sonication buffer (50 mM Tris-HCl, pH 8.0, 1 mM edetic acid, 300 mM NaCl, 1 mM dithiothreitol (DDT) , 1 mM PMSF) and sonicated on ice bath for 30 min. The supernatant was applied to a glutathione Sepharose 4B column equilibrated with Buffer A (50 mM Tris-HCl, pH 8.0, 1 mM DDT) and the column was washed with the same buffer. After the GST-fused SARS-E protein bound column was equilibrated with Buffer B (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10 mM reduced glutathione) at 4°C for 1 h, the eluate was collected. For SARS-3CL, N protein expression, bacterial cells was induced by IPTG (10 mM) at 22°C for 16h in tryptone-phosphate medium, proteins were extracted with buffer containing 50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 1 mM DTT, 1 mM Phenyl-methylsulfonyl fluoride, 10 mM imidazole, and 0.5 mg/ml lysozyme. The supernatant of the extract from 1 L of culture was loaded onto a Ni-NTA agarose column (8 ml) followed by washing with ∼100 ml of 20 mM imidazole in buffer A (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 1 mM DTT, and 1 mM Phenylmethylsulfonyl fluoride). Proteins were then eluted with 50 mM imidazole in buffer A.
Immunization and raising polyclonal antibodies
Rabbits were immunized s.c. at multiple sites on the back with 1 mg of indicated protein in 0.4 ml emulsion 1/1 v/v with complete Freund's adjuvant (CFA) containing 1 mg/ml Mycobacterium tuberculosis. The rabbits were boosted with the same antigens on 21 d and 42 d following the immunization. Blood was drawn from rabbits a week later following the second boost immunization and the blood was allowed to clot at 4°C. The antiserum was recovered by centrifugation.
Generation of monoclonal antibody against S2 fragment
Female BALB/c mice were immunized three times with the purified recombinant S2 protein. Spleen cells from the immunized mouse were fused with SP2/0 murine myeloma cells through standard procedures. Culture supernatants of the hybridoma were screened using indirect ELISA. After repeated screening and cloning with limiting dilution, four hybridoma clones were obtained. The mAb isotyping was carried out by using mouse sub-isotyping panel (Bio-Rad).
ELISA
The titers of the antisera from rabbits were determined by ELISA as described previously 14. In brief, 96-well (Corning Costar) were coated with the tested protein in 0.1M carbonate buffer (pH9.6) (20 mg/ml, 50 μl/well) at 4°C overnight. After blocking with PBS containing 10% fetal bovine serum (FBS) /0.1% Tween 20, and the plates were incubated with diluted rabbit antisera at various concentrations at 37°C for 2 h. Bound antibodies were determined with HRP-coupled goat anti-rabbit IgG (Bio-Rad) and the OD values at 450 nm were measured by microplate autoreader (Biotek).
SDS-PAGE and coomassie brilliant blue staining
Purified proteins were loaded in SDS-sample buffer [50 nM Tris-HCl (pH 6.8), 2% w/v SDS, 10% glycerol, 100 mM DTT and 0.1% w/v bromophennol blue] and size fractionated on 10% SDS-PAGE. The gel was stained by coomassie brilliant blue staining solution for 0.5 h and then destained.
Immunoprecipitation and western blotting
Purified proteins and SARS CoV infected Vero-E6 cell lysates were loaded in SDS-sample buffer [50 nM Tris-HCl (pH 6.8), 2% w/v SDS, 10% glycerol, 100 mM DTT and 0.1% w/v bromophennol blue] and size fractionated on SDS-PAGE (S1, S2, N protein on 10% SDS-PAGE, and E and 3CL protein on 15% SDS-PAGE). After being transferd to Nitrocellulose Transfer Membrane (pall corporation), the blots were incubated in blocking buffer (TBS containing 5% skim milk powder and 0.1% Tween 20) and probed with diluted rabbit antibody (1:10000) for 1 h. After washing, the membrane was incubated with HRP-conjugated goat anti-rabbit IgG (H+L) antibody (1:1500) (Southern Biotech), washed and analyzed using ECL reagents (Amersham Pharmacia Biotech).
For immunoprecipitiation, SARS-CoV (Beijing-01, see Supplement 1) infected Vero E6 cells were lysed in lysis buffer (20 mM sodium phosphate, pH 7.5, 500 mM NaCl, 0.1% SDS, 1% NP40, 0.5% sodium deoxycholate and 0.02% sodium azide with protease inhibitors) and then the lysate was centrifuge at 14,000 g in a precooled centrifuge for 15 min. The supernatant was then incubated with anti-S2 polyclonal antibody for 4 h in 4°C. Then appropriate amount of immobilized protein A was added to the antigen-antibody complex and incubated for further 2 h in 4°C. After 3 times wash, the beads were boiled for 5 min to dissociate the immuno-complexes from the beads. SDS-PAGE and western blot were performed and S2-specific mAb was used to detect the target protein.
Immuno-electron microscopic analysis
One drop of SARS-CoV culture supernatant was added onto the parafilm and then the carbon-coated copper mesh was put upside-down in it for 15 min at room temperature. The mesh was removed gently from the viral solution and the excess solution should be blotted up by sterilized tissue paper. And then the mesh was absorbed with the viral particles upside-down in a drop of antibody (against S1 or S2 or the serum from SARS patient) on the parafilm for 1 h at 37°C in a humid box. After being washed with PBS for three times the mesh was put in the gold-SPA conjugate diluted (1:20) in PBS for 1 h at 37°°C, followed by four times wash with 1% BSA in PBS. The excess solution should be absorbed by sterilized tissue paper. The mesh was then put in 3% tungstenic phosphate for 1 min at room temperature. After the excess solution was soaked up by sterilized tissue paper and dried in the air the mesh was observed by electron microscopy for the gold-conjugated viruses.
RESULTS
Protein expression and purification
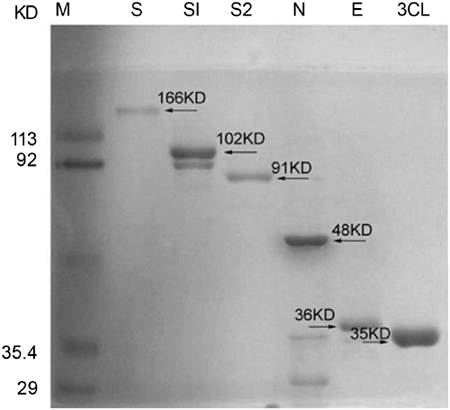
Based on bioinformatics analysis and available information, we supposed the cleavage site is between amino acid residues 667-668 7, therefore the S 1-667 and S 668-1255 were designed as S1 and S2 subunits, respectively. The full lengths of spike protein (S), and its subunits, S1 and S2, and small envelope (E) protein were expressed as GST-fusion proteins. Vector with His-tag was linked to the sequences of 3CL and nucleocapsid (N) genes. Most of S, S1 and S2 proteins were found in the insoluble fraction of the cell lysates, whereas N, E and 3CL proteins were found in the soluble fraction. The S, S1, S2 proteins were purified by Model 422 Electro-Eluter (Bio-rad) while E protein was purified by Glutathione Sepharose 4B (Amersham Biosciences) and 3CL proteinase and N protein were purified by Ni-column. As we expected, purified S1 protein (102 kD), S2 protein (91 kD), full length S protein (166 kD), N protein (48 kD), E-protein (36 kD) and 3-CL protein (35 kD) revealed clear bands at right positions on SDS-PAGE gel (Fig. 1).
Figure 1
Recombinant SARS-CoV nucleocapsid (N), spike (S, S1 and S2), small envelope (E) protein and 3-CL proteinase were purified and size fractionated on 10% SDS-PAGE. The gel was stained by coomassie brilliant blue staining.
Prokaryotic expressed proteins elicited high titer antibodies from immunized rabbits
After immunization with proteins, antisera against SARS-CoV N, S1 and S2, E and 3-CL proteins were collected. ELISA data showed that all the antisera had high affinity against the proteins used for immunization (Tab.1), suggesting that the expressed proteins of SARS-CoV were capable of inducing strong immune response in protein-immunized rabbits.
Table 1 Antibody titers against their corresponding proteins in protein-immunized rabbits were measured by ELISA. Before the immunization, sera were collected from rabbits (serve as pre-immunization) as control. Then the antisera were harvested following prokaryotic expressed proteins immunization in the rabbits one week later after last boost immunization (served as a post immunization).
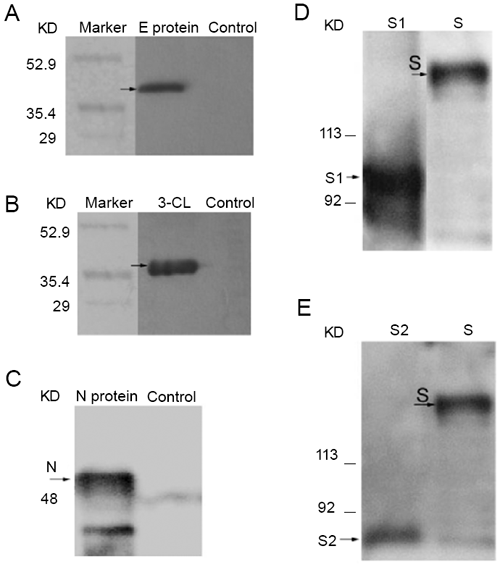
To test the binding activities of these antibodies, the expressed proteins were used to be served as targets and their reactions with antibodies were analyzed by western blot. The results showed that the antibodies against E and 3-CL, N proteins had strong and specific binding signals to their corresponding antigens (Fig. 2 A, B and C). It was previously expected that anti-S1 and S2 Abs should be able to bind to both the full-length of S protein and their corresponding fragments and the data did provide evidence for that hypothesis (Fig. 2 D and E). Our results suggested that both anti-S1 and S2 antibodies could be used for S protein detection.
Figure 2
Antibodies binding to their corresponding proteins by western blot analysis. (A) anti-E antibody; (B) anti-3CL antibody; (C) anti-N antibody; (D) anti-S1 antibody; (E) anti-S2 antibody. The proteins were subjected to SDS-PAGE and transferred to Nitrocellulose Transfer Membrane. The blots were probed with the antisera (1:10000) and detected with HRP-conjugated goat anti-rabbits IgG.
Anti-S1, S2 and N antibodies were able to bind to their corresponding proteins in the lysate of SARS-CoV infected Vero E6 cells
Since the antibodies were capable of binding to prokaryotic expressed proteins, we wondered whether the antibodies could also directly bind to corresponding viral proteins. To address this question, Vero E6 cells were infected by SARS-CoV for 24 h. Then the cell lysate were collected and western bolt was performed to determine the binding activity of these Abs. It showed that the antibodies against S1, S2 and N proteins could bind to their corresponding proteins and detect them (Fig. 3 A, B and C), while the antibodies against E and 3CL were not detecting their corresponding proteins (data not shown). The results demonstrated that anti-S1, S2 and N antibodies were specific for their corresponding proteins from SARS-CoV.
Figure 3
Antibodies binding to relevant proteins from the lysates of SARS-CoV infected Vero E6 cell. (A) anti-S1 antibody; (B) anti-S2 antibody; (C) anti-N antibody. The E.coli expressed S or N protein and the lysates were subjected to SDS-PAGE and transferred to Nitrocellulose Transfer Membrane. The blots were probed with the antisera (1:10000) and detected with HRP-conjugated goat anti-rabbits IgG.
Anti-S1 and S2 antibodies directly bound to the spike protein on the surface of SARS CoV
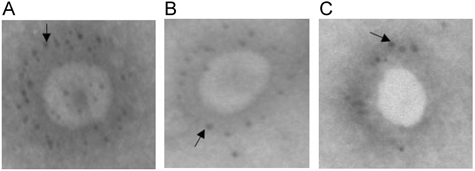
To evaluate whether anti-S1 and S2 Abs could directly bind to the virus, the binding activity of Abs to SARS-CoV viral particles were analyzed by electromicroscopy 15, 16. In order to give a positive control, serum containning high titer of antibodies against S protein (detected by ELISA-kit from BGL Life Tech Co.,Ltd, Beijing, China) was selected from a clinical diagnosed-SARS patient. The results showed that the complex of protein A-golden conjugate was capable of binding to the SARS-CoV viral particles in the presence of the serum of SARS patient (Fig. 4 A). Furthermore, the antibodies against S1 and S2 were used to detect the viral particles successfully (Fig. 4 B and C), suggesting that S1 and S2 Abs were able to bind to SARS-CoV.
Figure 4
Electromicroscopy of SARS-CoV with human Serum of SARS patients and rabbits antisera of S1 and S2. (A) sera from SARS patient; (B) anti-S1 Ab; (C) anti-S2 Ab. After staining with sera, the SARS-CoVs were incubated with the complex of protein-A and golden-particles conjugate. Then the images were observed by electron microscopy.
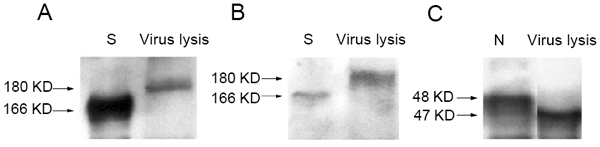
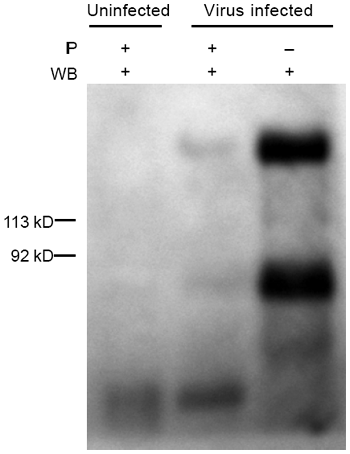
S2 fragment was detectable in the lysate of SARS-CoV infected Vero E6 cells
Given that S1 and S2 pAbs were both capable of binding to native S protein from the lysate and the S2 mAb is specific to detect S2 fragment, we assumed that if the S protein of SARS-CoV was cleaved during virus maturation in host cell, the S2 fragment should be detected by the S2 mAb in SARS-CoV infected Vero E6 cells. To address our questions, two experiments were designed. First, S2-specific mAb was directly used to detect native S2 fragment or full length of S protein in the lysate of SARS-CoV infected Vero E6 cells. Second, the lysate was initially immune-precipitated by S2 pAb and then the native S2 and S protein were detected by S2 mAb. The data showed that both S2 (about 85 kD) and S (180 kD) proteins were clearly detected in the two experimental conditions (Fig. 5), indicating the cleavage of S protein is present in SARS-CoV infected Vero E6 cells.
Figure 5
The S2-specifc monoclonal antibody to detect the subunit of S2 fragment. The lysate with or without immunoprecipitation (IP) with S2 pAb and then the target proteins were determined by S2 mAb using western blot (WB) analysis. The lysate was prepared from SARS-CoV infected Vero E6 cells.
DISCUSSION
SARS-CoV has brought damages to human health over the world. Till now little is known about this newly emerged virus. The antibodies would be helpful for us to study SARS-CoV. In this study SARS-CoV N protein, S, S1 and S2 proteins, E and 3-CL proteins were expressed in E. coli system and purified. The antibodies were raised from the immunized rabbits. Further studies proved that these antibodies provided a high affinity and specificity to their relevant antigens.
According to whether Spike protein is cleaved or not, coronaviruses can be divided into two groups. In the first group the spike glycoprotein is translated as large polypeptide which is cleaved into S1 and S2 fragments, and in the second the S protein is left untouched. The cleavage site within the first group remains relatively conservative. Studies in Bovine coronavirus, Avian infectious bronchitis virus, human coronavirus and mouse hepatitis viruses (MHVs) suggested that there is a conserved sequence between the S1 and S2 subunits and the motif of the sequence is RRSR 17, 20. However, we could not find such a motif in SARS-CoV. Based on bioinformatis analysis are deduced that the cleavage site was located in the amino acid residues between R667 and S668 (RS667–668). Here we subcloned the sequence of SARS-CoV spike gene as two subunits (S1 and S2) which were expressed respectively. The results demonstrated that both antibodies against S1 and S2 could bind to intact S protein, indicating that both antibodies will be useful for detecting S protein. Since the cleavage of S protein may be critical for the pathogenesis of SARS-CoV infectivity, the generated anti-S1 and S2 antibodies would allow us to study the interaction between S1 and S2 proteins and their functions.
Although the pAb can specifically bind to its corresponding protein, it may not exclude the possibility of the cross reaction with non-specific protein. In order to overcome this barrier, we also generated S2-speciflc mAb. It showed that both S2 and S proteins could be detected in the lysate, indicating that the cleavage of S protein may be involved in the SARS-CoV maturation. This finding will be helpful to explore the pathogenesis of SARS-CoV infectivity as well as to develop new drug for disease treatment. For example, assuming the cleavage was very important for the SRAS-CoV infection, the inhibition of this processes may consequently stop the virus infection. Our finding is consistent with the results from Dr. Marcus Eickmann's group, which also demonstrated that the S protein of SRAS-CoV was cleaved into S1 and S2 fragments in Vero E6 cells (personal communication).
Taken together, we have generated antibodies against some of the most important proteins of SARS-CoV and among these, antibodies against S1, S2 and N protein had relatively high specificity and affinity of binding to SARS-CoV. By using these antibodies, we demonstrated that the S protein underwent cleavage in Vero E6 host cells. We will further confirm the above findings in vivo or in other cell lines and investigate how this cleavage is involved in the SARS-CoV pathogenesis.
References
- Drosten C, Gunther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 2003; 348: 1967–76.
Article CAS PubMed Google Scholar - Ksiazek TG, Goldsmith CS, Zaki SR, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 2003; 348: 1947–59.
Article Google Scholar - Lipsitch M . Transmission dynamics and control of severe acute respiratory syndrome. Science 2003; 300: 1966–70.
Article CAS PubMed PubMed Central Google Scholar - Marra MA . The genome sequence of the SARS-associated coronavirus. Science 2003; 300: 1399–404.
Article CAS PubMed Google Scholar - Holmes KV, Enjuanes L . Perspective: The SARS coronavirus: A postgenomic era. Science 2003; 300: 1377–8.
Article CAS PubMed Google Scholar - Kuo L, Godeke GJ, Raamsman MJ, Masters PS, Rottier PJ . Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J Virol 2000; 74: 1393–406.
Article CAS PubMed PubMed Central Google Scholar - Gallagher TM, Buchmeier MJ . Coronavirus Spike Proteins in viral entry and pathogenesis. J Virol 2001; 279: 371–4.
Article CAS Google Scholar - Kuo L, Masters PS . Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J Virol 2002; 76: 4987–99.
Article CAS PubMed PubMed Central Google Scholar - Hiscox JA, Wurm T, Wilson L, et al. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J Virol 2001; 75: 506–12.
Article CAS PubMed PubMed Central Google Scholar - Tung FY, Abraham S, Sethna M, et al. The 9-kD hydrophobic protein encoded at the 3′end of the porcine transmissible gastroenteritis coronavirus genome is membrane-associated. Virology 1992; 186: 676–83.
Article CAS PubMed Google Scholar - Vennema H, Godeke GJ, et al. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J 1996; 15: 2020–8.
Article CAS PubMed PubMed Central Google Scholar - An S, Chen CJ, Yu X, et al. Induction of apoptosis in murine coronavirus-infected cultured cells and demonstration of E protein as an apoptosis inducer. J Virol 1999; 73: 7853–9.
CAS PubMed PubMed Central Google Scholar - Ziebuhr J, Herold J, Siddell SG, et al. Characterization of a human coronavirus (strain 229E) 3C-like proteinase activity. J Virol 1995; 69 :4331–8.
CAS PubMed PubMed Central Google Scholar - Lin Y, Shen X, Yang RF, et al. Identification of an epitope of SARS-coronavirus nucleocapsid protein. Cell Res. 2003; 13: 141–5.
Article CAS PubMed Google Scholar - Llames L, Goyache J, Domenech A, et al. Cellular distribution of bovine leukemia virus proteins gp51SU, Pr72(env), and Pr66(gag-pro) in persistently infected cells. Virus Res 1999; 79: 47–57.
Article Google Scholar - Nermut MV, Wallengren KK, Pager J J . Localization of actin in Moloney murine leukemia virus by immunoelectron microscopy. Virology 1999; 260: 23–34.
Article CAS PubMed Google Scholar - Mounir S, Talbot PJ . Molecular characterization of the S protein gene of human coronavirus OC43. J Gen Virol 1993; 74: 1981–7.
Article CAS PubMed Google Scholar - Abraham S, Kienzle TE, Lapps W, Brian DA . Deduced sequence of the bovine coronavirus spike protein and identification of the internal proteolytic cleavage site. Virology 1990; 176: 296–301.
Article CAS PubMed Google Scholar - Cavanagh D, Davis PJ, Pappin DJ, et al. Coronavirus IBV: partial amino terminal sequencing of spike polypeptide S2 identifies the sequence Arg-Arg-Phe-Arg-Arg at the cleavage site of the spike precursor propolypeptide of IBV strains Beaudette and M41. Virus Res 1986; 4: 133–43.
Article CAS PubMed PubMed Central Google Scholar - Schmidt I, Skinner M, Siddell S . Nucleotide sequence of the gene encoding the surface projection glycoprotein of coronavirus MHV-JHM. J Gen Virol 1987; 68: 47–56.
Article CAS PubMed Google Scholar
Acknowledgements
This work was supported by the grant of Technology Commission of Shanghai Municipality (No. 04DZ19108, 03DZ19113 and 03DZ19119), National Key Basic Research Program of China (No. 2001CB510006, 2002CB513008, 2003CB514117 and 2003CB514106), National Natural Science Foundations of China (No. 30170888, 30340032 and 30340031), Major State Basic Research Development program of China (No. G1999053907), SIPPE Foundation and the grant of Chinese Academy of Sciences (KSCX2-SW-225), grant of Sino-Germany center on SARS project [GZ238(202/11)], and grant from E-institutes of Shanghai Universities Immunology Division.
Author information
Author notes
- Xiao Dong WU, Bo SHANG, Rui Fu YANG, Hao YU, Zhi Hai MA and Xu SHEN: These authors contributed equally to this work.
Authors and Affiliations
- Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, 320 Yueyang Road, Shanghai, 200031, China
Xiao Dong WU, Bo SHANG, Zhi Hai MA, Yong Yong JI, Ying LIN, Ya Di WU, Guo Mei LIN, Lin TIAN, Xiao Qing GAN, You Hua XIE, Xue Liang ZHU, Gang PEI, Lin LI, Jia Rui WU & Bing SUN - Institute of Microbiology and Epidemiology, Institute of Basic Medical Sciences, Academy of Military Medical Sciences, Beijing, 100071, China
Rui Fu YANG, Sheng YANG & Wei Hong JIANG - Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, 300 Fenglin Road, Shanghai, 200031, China
Hao YU, Xu SHEN & Hua Liang JIANG - Institute of Materia Medica, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, 320 Taiyuan Road, Shanghai, 200031, China
Er Hei DAI & Xiao Yi WANG
Authors
- Xiao Dong WU
You can also search for this author inPubMed Google Scholar - Bo SHANG
You can also search for this author inPubMed Google Scholar - Rui Fu YANG
You can also search for this author inPubMed Google Scholar - Hao YU
You can also search for this author inPubMed Google Scholar - Zhi Hai MA
You can also search for this author inPubMed Google Scholar - Xu SHEN
You can also search for this author inPubMed Google Scholar - Yong Yong JI
You can also search for this author inPubMed Google Scholar - Ying LIN
You can also search for this author inPubMed Google Scholar - Ya Di WU
You can also search for this author inPubMed Google Scholar - Guo Mei LIN
You can also search for this author inPubMed Google Scholar - Lin TIAN
You can also search for this author inPubMed Google Scholar - Xiao Qing GAN
You can also search for this author inPubMed Google Scholar - Sheng YANG
You can also search for this author inPubMed Google Scholar - Wei Hong JIANG
You can also search for this author inPubMed Google Scholar - Er Hei DAI
You can also search for this author inPubMed Google Scholar - Xiao Yi WANG
You can also search for this author inPubMed Google Scholar - Hua Liang JIANG
You can also search for this author inPubMed Google Scholar - You Hua XIE
You can also search for this author inPubMed Google Scholar - Xue Liang ZHU
You can also search for this author inPubMed Google Scholar - Gang PEI
You can also search for this author inPubMed Google Scholar - Lin LI
You can also search for this author inPubMed Google Scholar - Jia Rui WU
You can also search for this author inPubMed Google Scholar - Bing SUN
You can also search for this author inPubMed Google Scholar
Corresponding authors
Correspondence toJia Rui WU or Bing SUN.
Rights and permissions
About this article
Cite this article
WU, X., SHANG, B., YANG, R. et al. The spike protein of severe acute respiratory syndrome (SARS) is cleaved in virus infected Vero-E6 cells.Cell Res 14, 400–406 (2004). https://doi.org/10.1038/sj.cr.7290240
- Received: 20 July 2004
- Revised: 20 August 2004
- Accepted: 24 August 2004
- Issue Date: 01 October 2004
- DOI: https://doi.org/10.1038/sj.cr.7290240