Emery-Dreifuss muscular dystrophy (original) (raw)
Clinical definition
Emery-Dreifuss muscular dystrophy (EDMD) is a relatively benign form of dystrophy, with onset in early childhood and thereafter relatively slow progression that is characterised by the triad:1
- 1
Early contractures, often before there is any significant weakness, of elbows, Achilles tendons, and post-cervical muscles (with subsequent limitation of neck flexion, but later forward flexion of the entire spine becomes limited) - 2
Slowly progressive muscle wasting and weakness with a distinctive humero-peroneal distribution (i.e. proximal in the upper limbs and distal in the lower limbs) early in the course of the disease. However weakness later extends to the proximal limb girdle musculature. Weakness is rarely profound - 3
Cardiac conduction defects (ranging from sinus bradycardia, prolongation of the PR interval on electrocardiography to complete heart block). A generalised cardiomyopathy may also supervene. Thus, affected individuals may die suddenly from heart block, or develop progressive cardiac failure. The latter may occur subsequent to the insertion of a pacemaker to correct an arrhythmia
Cardiac involvement is the most serious and important aspect of the disease. It usually becomes evident as muscle weakness progresses, but may exceptionally occur before there is any significant weakness. In almost all those affected by the disorder there is some evidence of cardiac involvement by age 30 years.
Two main modes of inheritance exist: X-linked and autosomal dominant. Rare autosomal recessive inheritance has also been described. Most X-linked EDMD patients become symptomatic in early childhood (<15 years) with mild weakness, followed by contractures. The disease is usually progressing and has a moderately benign course; loss of ambulation is exceptional. The disease course of the AD-EDMD is generally slow, but we can observe milder phenotype characterised by late onset and a mild degree of weakness and contractures or more severe phenotype with early presentation and a rapidly progressive course in few cases.2 It seems to be somewhat more severe than X-linked EDMD. A marked inter- and intra-familial variability in the clinical expression exists in patients with AD-EDMD.2
Diagnosis
Diagnosis is based just on clinical observation because there is no characteristic distinction in muscle biopsies. Immunohistochemistry of muscle biopsy tissue, leucocytes, fibroblasts or exfoliative buccal cells for emerin (X-linked EDMD), can confirm the diagnosis.3
Disease frequency
The disease frequency is estimated at 1/100 000 for the X-linked form but is unknown for the autosomal dominant inheritance. Only a few rare autosomal recessive cases have been described.4 There is a big proportion of sporadic cases with AD-EDMD.2
Gene
X-linked EDMD form
The gene responsible for the X-linked form was identified in 1994. It is located on chromosome Xq28. The STA gene is 2100 bp in length, consists in six exons and encodes 762 bp mRNA. Its 34 kD protein product of 254 amino acids has been designated ‘emerin’.5 Rat and mouse emerin sequences show >70% identity with human sequence, but there are no shared sequences in the drosophila melanogaster or C. elegans databases, except for the LEM domain which is also present in mammalian LAP2 and MAN1 proteins.6
Autosomal dominant EDMD form
The gene responsible for the autosomal dominant form was identified in 1999.7 It is LMNA and located on chromosome 1q11-q23. This gene spans approximately 24 kb and is composed of 12 exons. Alternative splicing within exon 10 gives rise to two different mRNAs: a 1992 pb mRNA that codes for pre-lamin A and a 1716 bp mRNA that codes for lamin C. Consequently, two proteins are generated, lamin A (664 aa, 74 kDa) and lamin C (572 aa, 64 kDa).8 Only the pre-lamin A, can be modified by isoprenylation. This protein cannot properly assemble into the nuclear lamina if it is not farnesylated and proteolytically processed to lamin A. In the mouse Lmna gene two separate promoters are identified, there is a somatic cell-acting promoter (for lamins A and C) and a testis-specific promoter (for lamin C2), which resides in the first intron. The presence of this alternative exon 1C2 that lead to lamin C2, has not been examined in humans so far, otherwise the genomic structure of murin and human LMNA genes is very similar. In both species the positions of intron insertion are exactly conserved, and the length of introns are also very similar.9
Autosomal recessive EDMD form
The implication of LMNA as the gene also responsible of the autosomal recessive form of the disease was described in 2000.4
Genetic heterogeneity
There is no other gene yet identified for which mutations are responsible of EDMD. However, it is important to note that mutations in LMNA gene were identified for three other autosomal dominant pathologies. Two of them are as EDMD, diseases of the striated muscles: the limb girdle muscular dystrophy type 1B (LGMD1B) and the dilated cardiomyopathy with conduction defects (DCM-CD).10,11,12 The third pathology is the Dunnigan type familial partial lipodystrophy (FPLD) affecting the adipose tissue.13 Two families have been described in which a LMNA mutation led to different phenotypes, EDMD, LGMD1B and DCM-CD.12,14 So far, no overlap between the three striated muscle disorders and FLPD have been described.
Function of the proteins
Emerin protein
Emerin is an inner nuclear membrane protein anchored to the inner nuclear membrane in skeletal, cardiac and smooth muscle, via a carboxy-terminal tail with remaining of the molecule projecting within the nucleoplasm. This protein presents several serine protein kinase sites.15 Emerin appears to be important in the organisation of the nuclear membrane during cell division. Fairley et al16 suggest that the primary roles of the emerin-nuclear protein complex is to stabilise the nuclear membrane against the mechanical stresses that are generated in muscle cells during contraction. Direct interaction between emerin and lamin A has been demonstrated by biomolecular interaction analysis on a BIAcore sensor and by immunoprecipitation analysis.17,18
Lamin A/C proteins
Lamins are components of the nuclear envelope and are located in the lamina, a multimeric structure associated with the nucleoplasmic surface of the inner nuclear membrane. Lamins A and C are members of the type V intermediate filament superfamily and are composed of different structural domains, an amino-terminal head, an α-helical rod domain made of heptade repeats, and a globular carboxy-terminal tail. Lamins form dimers through their rod domain and interact with the chromatin and integral proteins of the inner nuclear membrane (lamin B receptor, Lamin Associated Proteins (LAPs), emerin, Narf) through binding sites located in their rod domain and their carboxyl-terminal globular tail. There is compelling evidence that lamins play a role in DNA replication, chromatin organisation, spatial arrangement of nuclear pore complexes, nuclear growth, mechanical stabilisation of the nucleus and anchorage of the nuclear envelope proteins.8
Animal model
X-linked EDMD form
To date, there is no X-linked EDMD model reported.
Autosomal EDMD forms
Recently, Sullivan et al19 reported the derivation of mice in which the lamins A/C have been eliminated by gene targeting (by deletion of a region extended from exon 8 to the middle of exon 11), to produce either homozygous or heterozygous offspring. Both mice develop to term with no overt abnormalities. However, the post-natal growth of the homozygous mice is severely retarded, is characterised by appearance of muscular dystrophy. They exhibited premature mortality. This phenotype −/− is associated with ultrastructural perturbations to the nuclear envelope and with the mislocalisation of emerin in skeletal muscle. Mice heterozygous for the lmna mutation are overtly normal at 6–10 months with minimal evidence of dystrophy.
Mutations
STA gene
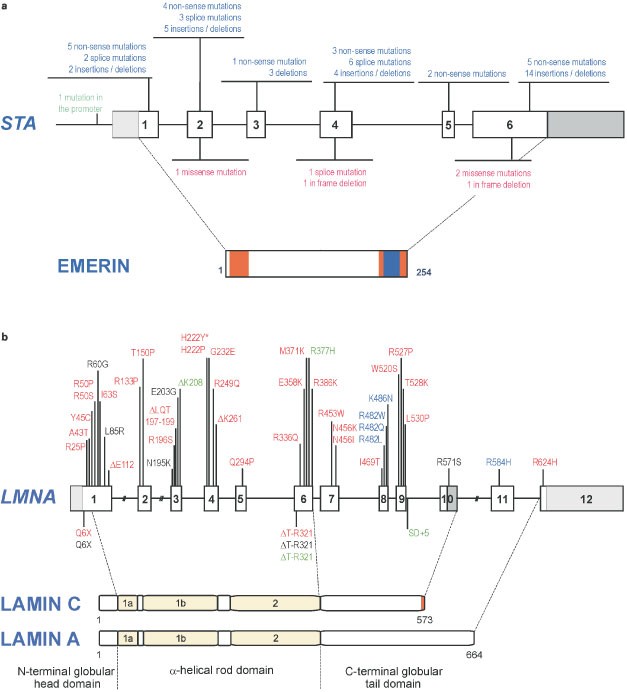
To date, around 100 mutations in the STA gene have been reported. They are approximately composed of 39.5% of small deletions, 31% of non-sense mutations, 15.5% of mutations in splice sites, 4% of large deletion of a part or the totality of the gene, 8.5% of missense mutations and 1.5% of mutation in the promoter (data base: www.path.cam.ac.uk/emd/mutation.html) (Figure 1a).20 Almost all mutations (86%) result in a complete absence of emerin on both Western blotting and immunocytochemistry. Since emerin mRNA levels are usually normal, absence of emerin suggests that truncated emerin mutants which lack the C-terminal transmembrane sequence are unstable under normal conditions. This enables rapid diagnosis of most X-linked cases by immunocytochemistry on skin biopsies or buccal smears or by Western blotting of white blood cells.3 Rare cases with reduced amount of the protein (due to a missense mutation) may have a milder phenotype.21 Ultrastructure analysis was performed on patient tissue: nuclear changes were observed in nuclei of skeletal muscle and cultured fibroblasts. Different degree of abnormalities in the nuclei, ranging from marked condensation of the chromatin to complete damage of the nuclear component, were observed in 10–18% of the cells. The extrusion of nuclear chromatin into sarcoplasm as a consequence of the nuclear membrane disintegration was observed in numerous nuclei.22,23
Figure 1
(a) STA mutations identified in XL-EDMD and their consequences on the protein structure of emerin. Mutations identified in XL-EDMD are presented.20 Above the gene are mutations leading to truncated emerin: insertions/deletions, splice site mutations and nonsense point mutation suppressing ATG or introducing a stop codon (in blue). Below the gene are mutations leading to mutated emerin: in frame deletions, missense point mutations and splice donor site mutations (in red). Blue box on emerin structure corresponds to the transmembrane domain; orange boxes correspond to part of emerin that are homologous to LAP2 domains.5 (b) LMNA mutations identified in AD-EDMD and other pathologies (LGMD1B, DCM-CD, FPLD) and their position on the protein structure of lamins A and C. Mutations identified in AD-EDMD are presented in red. The AR-EDMD mutation is depicted by an asterisk. LGMD1B mutations are in green. Black mutations correspond to DCM-CD mutations. The FPLD mutations are presented in blue. Above the gene are mutations leading to mutated lamins. Below the gene are mutations leading to truncated lamins.
LMNA gene
To date, 32 different mutations in LMNA gene are published to be responsible of AD-EDMD: 1 non-sense, 27 missense, 2 deletions of a codon, 1 deletion of 3 codons and 1 deletion of one nucleotide leading a frameshift (Figure 1b).2,4,24,25 They are distributed along the gene between exons 1 and 9 in the region common to lamins A and C excepted for one missense mutation which is located in exon 11 specific for lamin A.25 The mutation R453W was found in 16% of AD-EDMD patients. One mutation, H222Y, was identified at homozygous state in a patient from a consanguineous family. The unaffected parents carried the mutation at heterozygous state, demonstrating that LMNA mutations are responsible for the AR-EDMD.4 Mutations in LMNA gene are found in 100% of familial cases and in 35% of sporadic cases. The clinical picture is often compatible with other muscular dystrophies such as congenital muscular dystrophy or Bethlem muscular dystrophy. This could explain the low efficiency of LMNA mutation detection in the sporadic cases.
For the three striated muscles disorders (EDMD, LGMD1B and DCM-CD), there is no clear correlation between the phenotype and type or localisation of the mutation in the gene and a wide intra- and inter-familial clinical variability is observed (Figure 1b). For example, the Q6stop mutation identified in a big French family gives rise to phenotypes ranging from an isolated cardiac involvement, ie DCM-CD, to severe muscular and cardiac involvements.2,14 Another mutation, R366Q, was identified in a family in which only two out of the four members carrying the mutation, were affected.4 Finally, one severely affected patient possessed two mutations, one specific to lamin A that may modify the phenotype of this patient.25 Further studies are needed to identify the factors modifying striated muscle phenotypes among patients harbouring mutations within lamin A/C. In contrast, LMNA mutations described in FPLD affect for the majority of them (90%) the same codon of exon 8, R482.26
Immunocytochemical analysis of lamin A/C and emerin on skeletal muscle biopsies of AD-EDMD patients carrying LMNA mutation showed no detectable differences from control muscles, indicating that the mutations do not significantly alter the structure of the nuclear envelope.27 At ultrastructure level, nuclear changes were observed with alterations in the chromatin distribution. However, it is not in a large extent and only a small proportion of muscle nuclei exhibit abnormalities.27,28 Additional cases are needed before drawing any conclusion.
Treatment
There is no specific treatment. Development of specific therapeutic approaches may require a faithful animal model.
Great care should be given to proper diagnosis and follow-up of patients with EDMD. All patients should have a detailed cardiac investigation and regular follow-up by cardiologist since sudden death can be seen in these patients and early detection of arrhythmias can be lifesaving by pacemaker or defibrillator implantation. Also their relatives should be screened from the cardiologic point of view even if they show no subjective neuromuscular or cardiac symptoms. Apart from the patients with a typical presentation of weakness, contractures, and rhythm, especially in familial cases with a history of sudden death LMNA mutation should be searched for.
Accession numbers
(XL-EDMD, MIM no. 310300, STA gene coding for emerin); (AD-EDMD, MIM no. 181350, LMNA gene coding for lamin A/C).
References
- Emery AEH . Emery-Dreifuss muscular dystrophy – a 40 year retrospective Neuromusc Disord 2000 10: 228–232
Article CAS Google Scholar - Bonne G, Mercuri E, Muchir A et al. Clinical and molecular genetic spectrum of autosomal dominant Emery Dreifuss muscular dystrophy due to mutations of the lamin A/C gene Ann Neurol 2000 48: 170–180
Article CAS Google Scholar - Sabatelli P, Squarzoni S, Petrini S et al. Oral exfoliative cytology for the non-invasive diagnosis in X-linked Emery-Dreifuss muscular dystrophy patients and carriers Neuromusc Disord 1998 8: 67–71
Article CAS Google Scholar - di Barletta MR, Ricci E, Galluzzi G et al. Different mutations in the LMNA gene cause autosomal dominant and autosomal recessive Emery-Dreifuss muscular dystrophy Am J Hum Genet 2000 66: 1407–1412
Article CAS Google Scholar - Bione S, Maestrini E, Rivella S et al. Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy Nature Genet 1994 8: 323–327
Article CAS Google Scholar - Morris GE, Manilal S . Heart to heart: from nuclear proteins to Emery-Dreifuss muscular dystrophy Hum Mol Genet 1999 8: 1847–1851
Article CAS Google Scholar - Bonne G, Di Barletta MR, Varnous S et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy Nature Genet 1999 21: 285–288
Article CAS Google Scholar - Gruenbaum Y, Wilson KL, Harel A, Goldberg M, Cohen M . Review: nuclear lamins-structural proteins with fundamental functions J Struct Biol 2000 129: 313–323
Article CAS Google Scholar - Nakajima N, Abe K . Genomic structure of the mouse A-type lamin gene locus encoding somatic and germ cell-specific lamins FEBS Lett 1995 356: 108–114
Article Google Scholar - Muchir A, Bonne G, van der Kooi AJ et al. Identification of mutations in the gene encoding lamins A/C in autosomal dominant limb girdle muscular dystrophy with atrioventricular conduction disturbances (LGMD1B) Hum Mol Genet 2000 9: 1453–1459
Article CAS Google Scholar - Fatkin D, MacRae C, Sasaki T et al. Missense Mutations in the Rod Domain of the Lamin A/C Gene as Causes of Dilated Cardiomyopathy and Conduction-System Disease N Engl J Med 1999 341: 1715–1724
Article CAS Google Scholar - Brodsky GL, Muntoni F, Miocic S, Sinagra G, Sewry C, Mestroni L . Lamin A/C gene mutation associated with dilated cardiomyopathy with variable skeletal muscle involvement Circulation 2000 101: 473–476
Article CAS Google Scholar - Shackleton S, Lloyd DJ, Jackson SN et al. LMNA, encoding lamin A/C, is mutated in partial lipodystrophy Nature Genet 2000 24: 153–156
Article CAS Google Scholar - Bécane H-M, Bonne G, Varnous S et al. High incidence of sudden death of conduction system and myocardial disease due to lamins A/C gene mutation Pacing Clin Electrophysiol 2000 23: 1661–1666
Article Google Scholar - Nagano A, Koga R, Ogawa M et al. Emerin deficiency at the nuclear membrane in patients with Emery-Dreifuss muscular dystrophy Nature Genet 1996 12: 254–259
Article CAS Google Scholar - Fairley EA, Kendrick-Jones J, Ellis JA . The Emery-Dreifuss muscular dystrophy phenotype arises from aberrant targeting and binding of emerin at the inner nuclear membrane J Cell Sci 1999 112: 2571–2582
CAS PubMed Google Scholar - Sakaki M, Koike H, Takahashi N et al. Interaction between emerin and nuclear lamins J Biochem 2001 129: 321–327
Article CAS Google Scholar - Clements L, Manilal S, Love DR, Morris GE . Direct interaction between emerin and lamin A Bioch Biophys Res Com 2000 267: 709–714
Article CAS Google Scholar - Sullivan T, Escalante-Alcalde D, Bhatt H et al. Loss of A-type Lamin Expression Compromises Nuclear Envelope Integrity Leading to Muscular Dystrophy J Cell Biol 1999 147: 913–920
Article CAS Google Scholar - Yates JR, Wehnert M . The Emery-Dreifuss Muscular Dystrophy Mutation Database Neuromuscul Disord 1999 9: 199
Article CAS Google Scholar - Ellis J, Yates J, Kendrick-Jones J, Brown C . Changes at P183 of emerin weaken its protein-protein interactions resulting in X-linked Emery-Dreifuss muscular dystrophy Hum Genet 1999 104: 262–263
Article CAS Google Scholar - Fidzianska A, Toniolo D, Hausmanowa-Petrusewicz I . Ultrastructural abnormality of sarcolemmal nuclei in Emery-Dreifuss muscular dystrophy (EDMD) J Neurol Sci 1998 159: 88–93
Article CAS Google Scholar - Ognibene A, Sabatelli P, Petrini S et al. Nuclear changes in a case of X-linked Emery-Dreifuss muscular dystrophy Muscle Nerve 1999 22: 864–869
Article CAS Google Scholar - Felice KJ, Schwartz RC, Brown CA, Leicher CR, Grunnet ML . Autosomal dominant Emery-Dreifuss dystrophy due to mutations in rod domain of the lamin A/C gene Neurology 2000 55: 275–280
Article CAS Google Scholar - Brown CA, Lanning RW, McKinney KQ et al. Novel and recurrent mutations in lamin A/C in patients with Emery- Dreifuss muscular dystrophy Am J Med Genet 2001 102: 359–367
Article CAS Google Scholar - Vigouroux C, Magré J, Vantyghem MC et al. Lamin A/C gene: sex-determined expression of mutations in Dunnigan-type Familial Partial Lipodystrophy and absence of coding mutations in congenital and acquired generalized lipoatrophy Diabetes 2000 49: 1958–1962
Article CAS Google Scholar - Sewry CA, Brown SC, Mercuri E et al. Skeletal muscle pathology in autosomal dominant Emery-Dreifuss muscular dystrophy with lamin A/C mutations Neuropathol Appl Neurobiol 2001 27: 281–290
Article CAS Google Scholar - Sabatelli P, Lattanzi G, Ognibene A et al. Nuclear alterations in autosomal-dominant emery-dreifuss muscular dystrophy Muscle Nerve 2001 24: 826–829
Article CAS Google Scholar
Author information
Authors and Affiliations
- Inserm U523, Institut de Myologie, GH Pitié-Salpétrière, Paris, France
Anne Helbling-Leclerc, Gisèle Bonne & Ketty Schwartz
Authors
- Anne Helbling-Leclerc
You can also search for this author inPubMed Google Scholar - Gisèle Bonne
You can also search for this author inPubMed Google Scholar - Ketty Schwartz
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toGisèle Bonne.
Rights and permissions
About this article
Cite this article
Helbling-Leclerc, A., Bonne, G. & Schwartz, K. Emery-Dreifuss muscular dystrophy.Eur J Hum Genet 10, 157–161 (2002). https://doi.org/10.1038/sj.ejhg.5200744
- Received: 12 July 2001
- Revised: 05 September 2001
- Accepted: 07 September 2001
- Published: 25 April 2002
- Issue Date: 01 March 2002
- DOI: https://doi.org/10.1038/sj.ejhg.5200744