Isolation and characterisation of GTF2IRD2, a novel fusion gene and member of the TFII-I family of transcription factors, deleted in Williams–Beuren syndrome (original) (raw)
Introduction
Williams–Beuren syndrome (WBS (MIM: #19050)) is a rare developmental disorder (1/20 000 live births) characterised by a number of striking physical and behavioural features.1 Individuals with WBS typically present with a distinctive dysmorphic face, mild growth retardation, supravalvular aortic stenosis (SVAS) and often infantile hypercalcemia.1 These physical traits are combined with an unusual pattern of mental abilities (Williams Syndrome Cognitive Profile, WSCP) encompassing both enhanced verbal abilities and limited visuospatial capabilities, with characteristic ‘friendly’ personality traits.2
WBS is the result of a chromosomal microdeletion (typically ∼1.5 Mb in size) at 7q11.23 thought to arise as a consequence of unequal crossing over between highly homologous low-copy repeat (LCR) sequences flanking the deleted region.3 The majority of WBS patients have breakpoints, which cluster within these LCRs, but some possess atypical smaller deletions and consequently display partial phenotypes.4 Haploinsufficiency for elastin (ELN), the first gene identified in this region, causes the SVAS phenotype,5 lending support to the theory that individual genes located within the microdeletion are responsible for certain aspects of this complex syndrome.
The roles of many of the genes located in the WBS critical region have yet to be defined and their contribution to the pathology of WBS remains unclear, but recent work has been directed towards two I-repeat containing genes located near the telomeric end of the deletion breakpoints, which are invariably deleted in patients described with ‘classic’ WBS.6, 7 GTF2I (or TFII-I) and GTF2IRD1 (or GTF3, MusTRD1, BEN, CREAM, WBSCR11) both contain multiple I-repeats that are novel domains of a helix-loop-helix-like structure. GTF2I contains both DNA and protein binding sites and is a multifunctional transcription factor that can bind enhancer (E-box) and core promoter elements (Inr).6 Similarly, GTF2IRD1 is also thought to have gene regulatory function through directed DNA interactions.7
In our ongoing effort to complete the WBS transcription map, we have identified a novel gene, GTF2IRD2, located within the WBS critical region on chromosome 7. The mouse orthologue (Gtf2ird2) has also been isolated and maps to the syntenic mouse WBS region on chromosome 5G. We propose GTF2IRD2 to be the latest member of the I-repeat containing family of proteins (TFII-I family), comprising GTF2IRD1 and GTF2I (or TFII-I). GTF2IRD2 is, however, unusual because it has a C-terminal CHARLIE8 transposon-like motif thought to be a consequence of a random in-frame insertion of a transposable element that has generated a fusion gene. Here, we describe the characteristics and putative function of GTF2IRD2 in relation to WBS and the impact such a transposon-like element may have in this region. The location of GTF2IRD2 at the telomeric end of the WBS breakpoint, combined with its structural similarities to other genes in the WBS critical region (GTF2I and GTF2IRD1) and the novel C-terminal transposon-like element suggests that it has the potential to play a role in the pathogenesis of WBS.
Materials and methods
All primers described are summarised in Table 1. References 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18 describe the bioinformatics tools utilised for the characterisation of GTF2IRD2.
Table 1 PCR primer sequences
cDNA isolation
A mouse Gtf2ird2 cDNA clone was isolated by screening a Stratagene mouse embryo cDNA library with a probe made from BAC 391O16 (gb:AF267747). Hybridisation conditions were carried out according to the manufacturer's instructions (Stratagene) and repeats were competed out with placental DNA at a 100-fold concentration excess. Positive cDNA clones were sequenced by fluorescent BigDye terminator cycle sequencing (V 2.0 kit Applied Biosystems) using T7 and T3 vector primers, and visualised on an ABI 373 sequencer.
GTF2IRD2 was isolated from an 18-week human foetal brain cDNA library (Gibco) screened with probes made from the mouse Gtf2ird2 cDNA. Hybridisation conditions were according to the manufacturer's instructions. Positive cDNA clones were sequenced using vector primers T7 and T3 as described above.
Northern blot analysis
Human and mouse Northern blots of poly(A)+ RNA (Clontech) were probed with PCR products amplified from G_TF2IRD2_/Gtf2ird2 cDNA clones. Primers (GTF2IRD2 NTR/CTR and Gtf2ird2 × 3F-4R/CTR) were designed to exclude the I-repeat regions and to avoid cross hybridisation with similar genes. Hybridisation conditions were according to the manufacturer's instructions
RT-PCR analysis
Total RNA (1 _μ_g) from each tissue was reverse transcribed using a first-strand cDNA synthesis kit (Promega). Primers designed from specific regions (GTF2IRD2 NTR/CTR and Gtf2ird2 × 3F-4R/CTR) were used to look for expression by RT-PCR of the cDNA. PCR conditions were 50 ng of cDNA, 10 pmol of each primer and 0.5 U Taq polymerase (BCL) in the manufacturer's buffer for a 20 _μ_l reaction. Cycling conditions were 2 min denaturation at 94°C, followed by 35 cycles of 94°C 1 min, 55°C 1 min, 72°C 1 min and 5 min extension at 72°C. The integrity of the RNA was assessed by PCR analysis of the human GAPDH or mouse Gapdh gene. All primers were cDNA specific and spanned exon–intron boundaries to avoid amplification of contaminating DNA.
Deletion mapping of WBS somatic cell hybrids by PCR
The study was approved by the institutional review board. Hybrid cell lines containing the deleted chromosome 7 homologues were prepared from four patients with ‘classic’ WBS phenotypes, by fusion of the patient's lymphoblastoid cells with mouse BW5147 cells as described previously.4 The deletion status of WBSCR genes (primer sequences in Table 1) was determined by PCR analysis of DNA from the somatic cell hybrids. Specific GTF2IRD2 primers (GTF2IRD2 I7/I1 GTF2IRD2β) were designed by incorporating specific nucleotide differences identified in the intron sequences. Primers for the CALN1 gene were included as an internal nondeleted PCR control. Negative controls included mouse genomic DNA. PCR conditions were 50 ng of DNA, 10 pmol of each primer and 0.5 U Taq polymerase (BCL) in the manufacturer's buffer, for a 20 _μ_l reaction. Cycling was at 2 min denaturation at 94°C, followed by 30 cycles of 94°C 1 min, 55°C 1 min, 72°C 1 min, with a final 5 min extension at 72°C.
Results
Mus musculus Gtf2ird2 structure and properties
Mouse Gtf2ird2 cDNA clones were isolated by screening a Stratagene mouse embryo cDNA library with the BAC 391O16 (AF267747). The largest cDNA was 3468 bp in size and gave a predicted ORF of 936 amino-acid residues from the first predicted methionine start codon (in exon 2) (sequence submitted to GenBank: AY014963). The genomic structure of Gtf2ird2 was defined using the mouse BAC 391O16 sequence (gb: AF267747) (see Table 2). Gtf2ird2 spans a genomic region of 33 940 bp and is composed of 16 exons ranging from 29 to 1573 bp in size. All the exon–intron junctions conform to the consensus splice sequences. Two polyadenylation signals were identified 107 and 412 bp downstream from the termination codon (TAG) located in exon 16. Repeat masking program analysis15 of the cDNA sequence showed it to be composed of 46.86% (1619 bp) interspersed repeats: 2.92% were SINE/Alus (bases 20–116) and 43.94% were MER1_type (CHARLIE8) DNA element (bases 1457–1693 and 1716–2996).
Table 2 Mus musculus Gtf2ird2 genomic structure showing exon/intron sizes and boundaries
FISH analysis using BAC 391O16 suggests that there is a single copy of Gtf2ird2 residing on mouse chromosome 5G (data not shown) situated in a 5′–3′ orientation 3618 bp proximal to the Ncf1 gene (calculated from genomic sequence gb: AF267747).
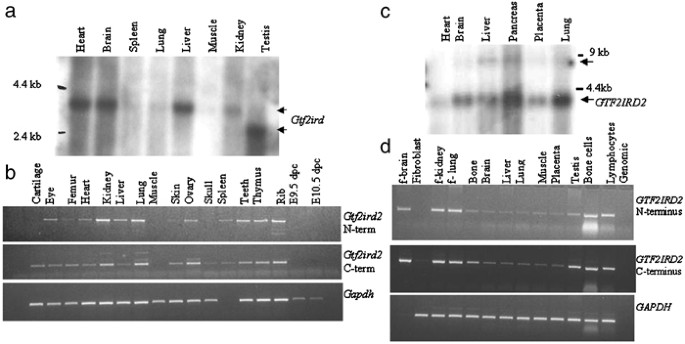
A Northern blot comprising adult mouse tissues hybridised with Gtf2ird2 cDNA probes displayed a strongly expressed transcript of approximately 3.5 kb in heart, brain and liver. Weaker bands were detected in spleen, lung, kidney and skeletal muscle. A smaller transcript (∼3 kb) was detected in testis suggesting that a splice isoform of Gtf2ird2 exists in this tissue. RT-PCR on a panel of mouse cDNAs using primers specific for the N- and C-termini showed that Gtf2ird2 was expressed in all the tissues tested, with significantly lower levels detected in heart, muscle, skin and bone (Figure 1a and b). There was no obvious expression in whole mouse embryo cDNA (E9.5 and 10.5 dpc).
Figure 1
Expression analysis of mouse and human GTF2IRD2. (a) Mouse Northern blot analysis showing a Gtf2ird2 transcript of ∼3.5 kb. A smaller transcript is present in mouse testis. (b) RT-PCR analysis shows Gtf2ird2 expression in all adult mouse tissues. (c) Human Northern blot analysis showing a GTF2IRD2 transcript of ∼3.5 kb in all tissues tested. (d) RT-PCR analysis shows ubiquitous GTF2IRD2 expression in all human tissues, but at very low levels in skin fibroblasts.
Human GTF2IRD2 structure and properties
The largest human GTF2IRD2 cDNA clone isolated from a human foetal brain cDNA library was 3559 bp in length with a predicted ORF 949 amino-acid residues (sequence submitted to GenBank: AY312850). Blast analysis of this cDNA sequence identified a number of genomic clones mapping to the WSCR with excellent homology, but only the sequence in BAC RP11-219M8 (gb: AC124781) was identical to the GTF2IRD2 cDNA we isolated. Sequence alignments allowed the genomic structure of GTF2IRD2 to be defined (see Table 3): it contains 16 exons comparable to those in the murine homologue, with all the intron/exon boundaries conforming to the consensus splice donor and acceptor sequences. Human GTF2IRD2 spans a genomic region of 57 251 bp and lies in a 3′–5′ orientation 6822 bp distal to the NCF1 gene. RepeatMasker15 analysis of the cDNA sequence identified a total of 2146 bp (60.30 %) of interspersed repeats. In all, 8.32% were ALUs, 1.46 % were low complexity repeats and 50.52% (1798 bp) was a MER1_type (CHARLIE8) DNA element located at the C-terminus.
Table 3 Human GTF2IRD2 genomic structure showing intron/exon sizes and boundaries
Northern blot analysis identified a GTF2IRD2 transcript of ∼3.5 kb with ubiquitous expression in the tissues tested (heart, brain, liver, pancreas, placenta and lung). This pattern was confirmed by RT-PCR analysis on a larger range of tissues (Figure 1). Higher levels of GTF2IRD2 appear to be expressed in foetal tissue compared with adult tissues with weak expression in skin fibroblasts.
Predicted GTF2IRD2 protein properties
GTF2IRD2 appears to be a slightly acidic protein (calculated pI of 5.8) with a molecular weight of 107 233 Da (936 Aa) and two discrete regions of predominantly hydrophilic residues (residues: 235–269 and 458–496) (Figure 2). Protein secondary structure analysis suggests a predominantly helical structure.18 It is also predicted to be soluble and located in the cytoplasm (77% probability).14 No signal peptide cleavage site was identified using the SignalP v1.1 program.16 Potential post-translational modifications include four N-glycosylation sites (residues: 156–159 NYSL; 204–207 NDSY; 465–468 NHSR; 552–555 NDTT) and multiple putative phosphorylation sites for all the major protein kinases including two tyrosine phosphorylation sites (residues: 235–243, 236–244).12, 14
Figure 2
GTF2IRD2β cDNA Sequence. Full-length sequence and annotated translation of GTF2IRD2β cDNA. Start is in bold text. The putative Leucine Zipper region is underlined with the residues implicated in bold text (VLLV). Two I-repeats are highlighted in grey, the CHARLIE8-like element (nucleotide 1434–3560) is in boldface. Predicted tyrosine phosphorylation sites (residues 243 and 244) are boxed. Amino-acid residues (478, 500, 597, 609, 612, 812) that differ in GTF2IRD2β and GTF2IRD2 are boxed.  indicates the BED zinc-finger DNA binding motif (at residue positions 435–471); the conserved aromatic and tryptophan motif (residues 418–429, 432) and the conserved cystine/histidine pattern (C_x_2C_x_ _n_H_x_3-5[H/C], residues 447–470). The atypical D,D(34)E domain residues (748, 782 and 816) are highlighted with an asterisk (*). No polyA signal was detected in the cDNA sequence.
indicates the BED zinc-finger DNA binding motif (at residue positions 435–471); the conserved aromatic and tryptophan motif (residues 418–429, 432) and the conserved cystine/histidine pattern (C_x_2C_x_ _n_H_x_3-5[H/C], residues 447–470). The atypical D,D(34)E domain residues (748, 782 and 816) are highlighted with an asterisk (*). No polyA signal was detected in the cDNA sequence.
The sequence divides into two halves: the N-terminal 410 amino acids show high homology to the TFII-I family of transcription factors (GTF2I and GTF2IRD1) and contain two I-repeats (PF02946, IPR004212) (residues: 107–185 and 332–410). The majority of the identity shared between GTF2IRD1 and GTF2IRD2 is located around the I-repeats with little conservation in the remaining sequence. The N-terminus of GTF2IRD2 (residues 1–410) has 75% identity and 84% similarity with GTF2I (see Figure 3). The C-terminal sequence (residues 414–949) of GTF2IRD2 share high identity (>78%) to the CHARLIE8 transposable element.
Figure 3
TFII-I family protein alignments. ClustalW (1.82) alignments of the N-terminal regions of the human I-repeat containing proteins (TFII-I family). The residues implicated in the putative leucine zipper motif are highlighted in grey and the I-repeats are boxed. Full-length GTF2I (NM_016328) is used in this alignment alongside GTF2IRD1 (NM_016328) and GTF2IRD2 (AY312850).
DNA and protein binding capabilities of GTF2IRD2
GTF2IRD2 has numerous putative sites thought to be capable of facilitating DNA and protein interactions. An N-terminal leucine zipper (LZ) motif, situated before the first I-repeat at amino-acid residues 23, 30, 37, 44 (VLLV), is thought to aid the formation of dimers.19 A BED zinc-finger DNA binding motif (PF02892) was identified at positions 435–471.11 The BED zinc-finger motif of 50–60 residues comprised of conserved cystines and histidines (C_x_2C_x_ _n_H_x_3–5[H/C]),20 and an N-terminal conserved tryptophan motif. In GTF2IRD2, the metal chelating motif is clearly visible (residues 447, 450, 465 and 470,) but the N-terminal, mainly aromatic, motif is slightly less well preserved with three (amino acids 432, 433, 435) of the five key residues conserved (Figure 3). A region of basic residues (KRK) N-terminal to the BED motif at positions 418–420 is thought to interact with the minor groove of DNA, while the rest of the BED structure associates with the major groove.20 In addition, GTF2IRD2 has a C-terminal CHARLIE8 transposon-like element of 534 residues. The CHARLIE8 transposon (or MER102)21 is an autonomous mammalian-specific member of the MER1 transposase family22 characterised by an 8 bp palindromic insertion site and two flanking Terminal Inverted Repeats (TIRs – 15 bp consensus sequence: CAGgGGTCCCCAACC).23 Most transposable elements are autonomous and transposition is catalysed by the transposase encoded by the element itself. The transposase functions by binding TIRs flanking the transposable element and then cleaving the DNA to excise the element, leaving a characteristic footprint behind (a 16 bp duplication of the 8 bp target site). To determine if the integrated CHARLIE8 transposon-like element could still be functional, GTF2IRD2 and the surrounding genomic sequence was screened for three transposase-related domains:
- i)
MER1 TIR sequences: two putative sites flanking the CHARLIE8-like element were identified – one in the genomic sequence ∼3922 bp upstream of GTF2IRD2 3′ UTR and one in intron 15 approximately 137 bp upstream from exon 16. - ii)
the ‘third region’ is a motif required for the transposase ability of members of the hAT transposase family;24 this motif was detected at amino-acid residues 884–949. - iii)
an atypical D,D(35)E domain (D,D(34)E), which is the DNA cleavage and excision active site;25 this motif was detected at residues 748, 782 and 816 with conserved spacing between the residues.
The genomic sequence between NCF1 3′UTR and GTF2IRD2 3′UTR was also analysed using exon prediction packages (MZEF, FEX and Hexon) to identify the rest of the original gene disrupted by the CHARLIE8-like transposase, however, no obvious partial transcripts were detected. The CHARLIE8 transposon-like motif appears to be present only on chromosome 7. Other putative fusion genes described with similar architecture to GTF2IRD2 (ie a cellular gene comprising of an N-terminal hAT transposase-like element) include: Buster1 (NP_067034) on chromosome 11p15.3 and Buster3 (NP_071373) on chromosome 5q34. Both genes have unknown functions but are thought to be ‘fusion genes’ created by random in-frame insertions of hAT transposase-like mobile elements.22
Similar analyses on the mouse Gtf2ird2 protein identified a BED finger motif (KRK) at residues 414–416 with the N-terminal, mainly aromatic, residues (WCCHH at residues 428, 442, 445, 460 and 465), and an atypical DDE domain (residues 742, 776 and 810). No ‘third region’ was detected.
Duplication of _GTF2IRD_2 in the human WBSCR
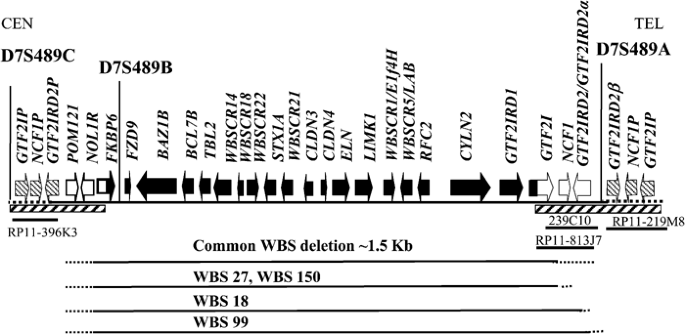
The deletion in WBS is flanked by low copy repeats3 and since GTF2IRD2 maps within this duplicated region, more than one copy of the gene was identified by database mining (predominantly BLAST searches). Alignment of all the GTF2IRD2 sequences together suggests that there could be three full-length transcript variants and a pseudogene, at different loci around the WBSCR (see Figure 4).
- i)
The GTF2IRD2 gene (gb: AY312853) resides in the genomic clones BAC 239C10 (gb: AC004166) and PAC RP4-771P4 (gb: AC004883)) and is supported by cDNA clone evidence (clone DKFZp434O1635 gb: AL834153). - ii)
GTF2IRD2α (gb: AY312854) lies in BAC RP11-813J7 (gb: AC083884) and is supported by a recently submitted cDNA clone (gb: NM_173537). - iii)
Our isolated cDNA clone resides in BAC RP11-219M8 (gb: AC124781), which subsequently influenced the naming of our gene to GTF2IRD2β (gb: AY312850) because it lies distal to the other GTF2IRD2 gene(s).
Figure 4
Transcript map of the WBS critical region showing the loci of the GTF2IRD2 genes and the extent of the deletions in some WBS individuals. Hatched boxes indicate duplicated regions. At the telomeric end, GTF2IRD2 and GTF2IRD2α probably represent the same gene copy but they differ by two base changes/polymorphisms. The precise location of the WBS patient breakpoints within the duplicated regions is unknown. The location of the genomic clones containing the GTF2IRD2 genes are shown and the region is in a centromeric to telomeric orientation.
The genomic clones containing GTF2IRD2 and GTF2IRD2α appear to be overlapping in the sequence contig (see Figure 4); however, GTF2IRD2α has two different amino-acid residues (K39E) and (N514H). These could be natural polymorphisms, in which case GTF2IRD2 and GTF2IRD2α would represent the same copy of the gene. In contrast, GTF2IRD2β contains six nucleotide differences that, in addition to its location, distinguishes it from GTF2IRD2. These sequence differences lie at the C-terminus of the sequence resulting in amino-acid changes at residues 478, 500, 597, 609, 612 and 812 (see Figure 3). None are truncating and do not affect the size of the predicted ORF, indicating that this copy may not be a pseudogene and is probably functional. In addition, like GTF2IRD2, putative TIRs were detected in the genomic region harbouring the GTF2IRD2β gene (one in intron 15–145 bp before exon 16 and one ∼3190 bp after the 3′UTR sequence). cDNA clones for all the GTF2IRD2 variants described were identified suggesting that they are all transcribed in vivo.
In addition, a putative pseudogene (GTF2IRD2P, gb: AY312852) was predicted from genomic clone RP11-396K3 (gb: AC006995) and maps to the centromeric duplicated region flanking the WBSCR. GTF2IRD2P is a partial gene (ORF of 578 amino-acid residues) with a number of sequence variants including a deletion that results in a frameshift and consequently a premature stop codon. Database mining did not uncover any cDNA clones for the pseudogene.
Genomes of other mammalian species, sequenced to date, that contain the GTF2IRD2 orthologous genes include: Bos taurus, Sus scrofa, Felis catus and Papio cynocephalus anubis. No Drosophila, C.elegans or Fugu homologues were detected, indicating that GTF2IRD2 appears to be a mammalian-specific gene.
Deletion status of GTF2IRD2 in WBS patients
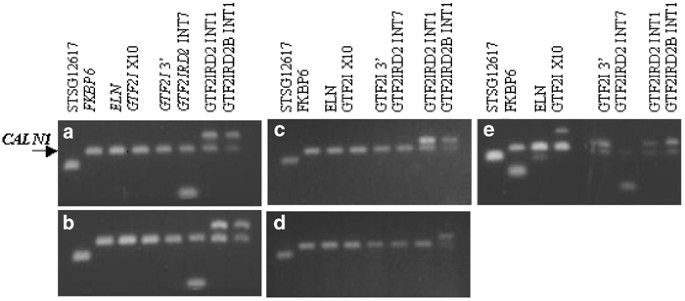
Somatic cell hybrid lines containing the deleted chromosome 7 homologues were prepared from four individuals with WBS and screened for the presence of the GTF2IRD2 gene by PCR analysis using locus-specific primers for intron 7 and intron 1, thereby, allowing selective amplification of GTF2IRD2. Primers that specifically amplify intron 1 of GTF2IRD2β were also used to determine the extent of the deletions. Two patients (WBS 27 and WBS 150) had deletions encompassing GTF2I and two (WBS 18 and WBS 99) had larger deletions extending to GTF2IRD2 (Figures 4 and 5). Clinical descriptions of the patients deleted for GTF2IRD2 did not immediately highlight any phenotypic differences from the nondeleted cases, and all were described with ‘classic’ WBS phenotypes.
Figure 5
Deletion mapping of somatic cell hybrids. PCR analysis of somatic cell hybrids from the deleted chromosome 7 homologues of WBS individuals alongside a normal control. (a) WBS 27, (b) WBS 150, (c) WBS 18, (d) WBS 99 and (e) normal control. CALN1 is an internal nondeleted PCR control. Marker STSG12617 lies ∼2 Mb proximal to the WBSCR and is not deleted in WBS.
Discussion
We have identified a novel fusion gene, GTF2IRD2, characterised structurally by the presence of two N-terminal I-repeats and a C-terminal CHARLIE8 transposon-like element.22 GTF2IRD2 resides in the telomeric duplicated region flanking the WBS critical region and has at least three potentially functional full-length variants mapping to loci close to each other on chromosome 7q11.23. The mouse orthologue shows 80% homology to the human protein but is present as a single copy on chromosome 5G.
GTF2IRD2 is the third member of the TFII-I family of transcription factors (GTF2I and GTF2IRD1) to be described.6 All members reside on 7q11.23 and are characterised structurally by the presence of multiple I-repeats, each containing a helix-loop-helix-like domain. The I-repeat is thought to be involved in DNA and/or protein interactions.7 When considering the overall biological role of GTF2IRD2, its shared homology to the TFII-I family suggests that it may act as a transcription factor regulated by particular signalling cascades.6 However, GTF2IRD2 differs from the other TFII-I members because it has a C-terminal CHARLIE8 transposon-like region, the role of which is as yet unclear. Generally, transposases offer no selective advantage to the host and act merely to insert and excise themselves randomly from the genome. If, however, a transposable element inserts into a gene rather than a noncoding sequence the effects can be profound. Often the gene will be adversely disrupted, but in rare cases the transposase can insert in-frame and produce a viable protein.26 Such fusion proteins will either be lost from the genome during evolution or, if the fusion product provides the protein with novel functionality, it will be retained and have a biological role.22 This may be the case with GTF2IRD2, where the CHARLIE8 transposon has inserted itself in-frame creating a fusion gene, which is probably functional since it is highly conserved in mammals.
Although the CHARLIE8 transposase itself is unlikely to be active as an individual protein, the CHARLIE8-like element present in GTF2IRD2 appears to have retained a number of domains and motifs intrinsically linked to transposase ability – (i) a region with significant identity to the BED DNA binding domain;20 (ii) an atypical D,D(35)E domain;20 considered to be a crucial active site whose disruption results in loss of transposase ability.25 Partially functioning atypical D,D(35)E domains have been observed in other proteins, for example, the Centromere protein B. CENP-B is a constitutive protein that has retained the ability to cleave single-stranded DNA on recognition of a highly conserved 17 bp motif located at most mammalian centromeres (CENP-B box), but it lacks the ability to subsequently excise the fragment.27 The cutting action of CENP-B on DNA in the vicinity of the CENP-B boxes is thought to destabilise the region, promoting homologous recombination in ‘hotspots’; (iii) a ‘third region motif’ essential for hAT transposase function24 and (iv) two putative TIR sequences flanking the region thought to be the transposase insertion site.
The presence of these motifs suggests that the CHARLIE8-like region of GTF2IRD2 may have retained limited transposase functionality. This may just be the ability to interact with selected DNA or protein motifs, or may be more wide-ranging extending to strand cleavage. Alternatively, their presence may allow other MER1 elements to bind and cleave, resulting in regional instability.28 The presence of transposase target sites influencing the stability of a genomic region has been proposed as the pathological mechanism in Charcot–Marie–Tooth neuropathy type 1A (CMT1A, [MIM: 118220]) and hereditary neuropathy with liability to pressure palsies (HNPP, [MIM: 162500]). Both conditions occur due to unequal crossover events between misaligned CMT1A-REP repeats flanking a 1.5 Mb region on chromosome 17.29 Duplications at CMT1A-REP repeats residing on 17p11.2-12 cause CMT1A, while the reciprocal deletions are associated with HNPP.30 In all, 76% of crossover events in CMT1A and HNPP occur at a recombination hotspot containing a _mariner_-like element (MITE) flanked by a 36 bp inverted repeat.30 Although MITE is not thought to encode a functional transposase, its presence is thought to influence the stability of the region by allowing other active transposases to bind and cleave the MITE TIR sequences. One can, therefore, envisage that the presence of transposase element target sites residing within the duplicated 7q11.23 regions harbouring the GTF2IRD2 genes could be a further factor influencing the instability of the region, by a similar mechanism to HNPP, making it more susceptible to gross chromosome rearrangements. Not all of these sites are conserved in the mouse Gtf2ird2 protein, indicating that it is less likely to have transposase activity in that species.
GTF2IRD2 is deleted in some WBS patients with the full spectrum of clinical phenotypes, but its role in the pathology of the disorder is not yet clear. Further studies will involve more detailed clinical and psychological analyses on these patients to determine whether any phenotypic differences exist that can be attributed to haploinsufficiency for this gene. In conclusion, GTF2IRD2 is a novel gene that could be involved in the pathology of WBS, and may also act as an element that undermines the stability of the critical region and, consequently, plays a role in promoting the unequal homologous recombination events underlying the WBS deletion.
References
- Morris C : The natural history of Williams syndrome: physical characteristics. J Paediatr 1988; 113: 318–326.
Article CAS Google Scholar - Pober B, Dykens E : Williams syndrome: an overview of medical, cognitive, and behavioural features. Child Adolesc Psychiatr Clin N Am 1996; 5: 929–943.
Article Google Scholar - Valero M, de Luis O, Cruces J, Pérez Jurado L : Fine-scale comparative mapping of the human 7q11.23 region and the orthologous region on mouse chromosome 5G: the low-copy repeats that flank the Williams–Beuren syndrome deletion arose at breakpoint sites of an evolutionary inversion(s). Genomics 2000; 69: 1–13.
Article CAS Google Scholar - Tassabehji M, Metcalfe K, Karmiloff-Smith A et al: Williams syndrome: use of chromosomal microdeletions as a tool to dissect cognitive and physical phenotypes. Am J Hum Genet 1999; 64: 118–125.
Article CAS Google Scholar - Ewart A, Morris C, Atkinson D et al: Hemizygosity at the elastin locus in a development disorder, Williams syndrome. Nat Genet 1993; 5: 11–16.
Article CAS Google Scholar - Roy A : Biochemistry and biology of the inducible multifunctional transcription factor TFII-I. Gene 2001; 274: 1–13.
Article CAS Google Scholar - Vullhorst D, Buonanno A : Characterisation of general transcription factor 3, a transcription factor involved in slow muscle-specific gene expression. J Biologic Chem 2003; 278: 8370–8379.
Article CAS Google Scholar - Rice P, Longden I, Bleasby A : EMBOSS: the European molecular biology open software suite. Trends Genet 2000; 16: 276–277.
Article CAS Google Scholar - Thompson J, Higgins D, Gibson T : CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 1994; 22: 4673–4680.
Article CAS Google Scholar - Wilkins M, Gasteiger E, Bairoch A et al: Protein identification and analysis tools in the ExPASy server; In: Link A (ed): 2-D proteome analysis protocols. New Jersey, Humana Press; 1998.
Google Scholar - Bateman A, Birney E, Cerrut L et al: The Pfam protein families database. Nucleic Acids Res 2002; 30: 276–280.
Article CAS Google Scholar - Gattiker A, Gasteiger E, Bairoch A : ScanProsite: a reference implementation of a PROSITE scanning tool. Appl Bioinform 2002; 1: 107–208.
CAS Google Scholar - Altschul S, Madden T, Schäffer A et al: Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997; 25: 3389–3402.
Article CAS Google Scholar - Nakai K, Kanehisa M : A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 1992; 14: 897–911.
Article CAS Google Scholar - Smit A, Green P RepeatMasker program, unpublished data.
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G : Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 1997; 10: 1–6.
Article CAS Google Scholar - Emanuelsson O, Nielsen H, Brunak S, von Heijne G : Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 2000; 300: 1005–1016.
Article CAS Google Scholar - Wilmot C, Thornton J : Beta-turns and their distortions: a proposed new nomenclature. Protein Eng 1990; 3: 479–493.
Article CAS Google Scholar - Ferré-D'Amar A, Prendergast G, Ziff E, Burley S : Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature 1993; 363: 38–45.
Article Google Scholar - Aravind L : The BED finger, a novel DNA-binding domain in chromatin-boundary-element-binding proteins and transposases. Trends Biologic Sci 2000; 25: 421–423.
Article CAS Google Scholar - Jurka J, Naik A, Kapitonov V : CHARLIE8 RepBase entry RepBase release 7.2. (Accessed 7th November 2002) 1998.
- Smit A : Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev 1999; 9: 657–663.
Article CAS Google Scholar - Smit A, Riggs A : Tiggers and other DNA transposon fossils in the human genome. Proc Natl Acad Sci USA 1996; 93: 1443–1448.
Article CAS Google Scholar - Calvi B, Hong T, Findley S, Gelbert W : Evidence for a common evolutionary origin of inverted repeat transposons in drosophila and plants; hobo, Activator, and Tam3. Cell 1991; 66: 465–471.
Article CAS Google Scholar - Kulkosky J, Jones K, Katz R, Mack J, Skalka A : Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol 1992; 12: 2331–2338.
Article CAS Google Scholar - Esposito T, Gianfrancesco F, Ciccodicola A et al: A novel pseudoautosomal human gene encodes a putative protein similar to Ac-like transposases. Hum Mol Genet 1999; 8: 61–67.
Article CAS Google Scholar - Kipling D, Warburton P : Centromeres, CENP-B and Tigger too. Trends Genet 1997; 13: 141–145.
Article CAS Google Scholar - McCarron M, Duttaroy A, Doughty G, Chovnick A : Drosophila P element transposase induces male recombination additively and without requirement for a P element excision or insertion. Genetics 1994; 136: 1013–1023.
CAS PubMed PubMed Central Google Scholar - Kiyosawa H, Chance P : Primate origin of the CMT1A-REP repeat and analysis of a putative transposon-associated recombinational hotspot. Hum Mol Genet 1996; 5: 745–753.
Article CAS Google Scholar - Reiter L, Murakami T, Koeuth T et al: A recombination hotspot responsible for two inherited peripheral neuropathies is located near a mariner transposon-like element. Nat Genet 1996; 12: 288–297.
Article CAS Google Scholar
Acknowledgements
This research was supported by the Wellcome Trust (Grant No. 061183).
Author information
Authors and Affiliations
- University of Manchester, Academic Unit of Medical Genetics and Regional Genetic Service, St Mary's Hospital, Manchester, UK
Hannah J Tipney, Timothy A Hinsley & May Tassabehji - Department of Computer Science, University of Manchester, Oxford Road, Manchester, UK
Andrew Brass - School of Biological Science, University of Manchester, Oxford Road, Manchester, UK
Timothy A Hinsley & Andrew Brass - Academic Unit of Medical Genetics and Regional Genetics Service, St Mary's Hospital, Manchester, UK
Kay Metcalfe & Dian Donnai
Authors
- Hannah J Tipney
You can also search for this author inPubMed Google Scholar - Timothy A Hinsley
You can also search for this author inPubMed Google Scholar - Andrew Brass
You can also search for this author inPubMed Google Scholar - Kay Metcalfe
You can also search for this author inPubMed Google Scholar - Dian Donnai
You can also search for this author inPubMed Google Scholar - May Tassabehji
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toMay Tassabehji.
Rights and permissions
About this article
Cite this article
Tipney, H., Hinsley, T., Brass, A. et al. Isolation and characterisation of GTF2IRD2, a novel fusion gene and member of the TFII-I family of transcription factors, deleted in Williams–Beuren syndrome.Eur J Hum Genet 12, 551–560 (2004). https://doi.org/10.1038/sj.ejhg.5201174
- Received: 26 September 2003
- Revised: 12 December 2003
- Accepted: 07 January 2004
- Published: 21 April 2004
- Issue Date: 01 July 2004
- DOI: https://doi.org/10.1038/sj.ejhg.5201174