Mutations in TBX1 genocopy the 22q11.2 deletion and duplication syndromes: a new susceptibility factor for mental retardation (original) (raw)
Introduction
The 22q11.2-deletion syndrome associated with DiGeorge or velocardiofacial syndromes (DGS/VCFS; MIM188400) is the most frequent microdeletional syndrome with an incidence of 1/4000 births. Although most patients carry the same 3 Mb hemizygous deletion they display a very variable phenotype that ranges from normality to isolated heart defects, to multiple defects that include, dysmorphic facial features, cardiac outflow abnormalities, absence or hypoplasia of the thymus and parathyroid gland, mental retardation, psychiatric problems and velopharyngeal insufficiency.1 In addition, the reverse genomic defect, 22q11.2 duplications, have been recently described to also cause very diverse phenotypes that range from normality to isolated mental retardation, to multiple defects similar to those found in the 22q11.2-deletion syndrome.2, 3, 4, 5, 6, 7
Over 30 genes map within the commonly deleted/duplicated 3 Mb region; however, several studies indicate that haploinsufficiency of TBX1 is responsible for many of the phenotypic traits of the 22q11.2-deletion syndrome.8, 9, 10 TBX1 codes a transcription factor that belongs to the T-box family and is expressed in early embryonic structures from which the affected organs originate. Mice nullyzygous for Tbx1 present similar features to those of DiGeorge patients, whereas mice that are heterozygous reproduce similar cardiac outflow abnormalities. In addition, TBX1 may also be responsible for the phenotypic traits associated with the 22q11.2 duplication, as mice carrying extra copies of TBX1 have been found to display the full spectrum of malformations of the 22q11.2-deletion syndrome.11
Up to now, only four families have been found to harbour heterozygous mutations in the TBX1 gene in individuals with features of DGS/VCFS syndrome lacking the deletion.12, 13 In the first study, Yagi et al13 detected three mutations that segregated with DGS or VCFS phenotypes, but none of the patients displayed mental retardation. In the second study by Paylor et al,12 a mutation was found in a family affected by VCFS and a member with an autistic spectrum disorder, Asperger syndrome. Other studies have found rare variants with uncertain clinical significance, as they are not present in the normal population but do not segregate with the phenotypic features of the syndrome within the family, or functional information is unavailable.14, 15, 16
To gain further insight on the role of TBX1 in human disease, we have screened 38 non-deleted patients who have some of the clinical features compatible with the 22q11.2-deletion syndrome.
Materials and methods
Patients
Patients were divided into three groups attending their phenotypic traits. Group 1: 23 patients with cardiac outflow tract abnormalities as the only clinical feature; group 2: five patients with other phenotypic features of the 22q11.2-deletion syndrome without heart involvement and group 3: 10 patients who had heart abnormalities together with at least another of the clinical features of the 22q11.2 syndrome. Blood samples were obtained from patients and parents after written informed consent. Typical and atypical deletions and duplications in the 22q11.2 region were ruled out in patients using a combination of different techniques: Fluorescent in situ hybridisation (FISH), multiple ligation probe assay (MLPA) and microsatellite typing.
Two hundred patients who had been referred because of unspecific mental retardation and/or hyperactive behaviour, which tested negative for the fragile-X syndrome, were screened for a specific 5′UTR TBX1 mutation. All of these patients had normal karyotypes and some of had been tested for other diseases, Prader–Willi or Angelman syndromes by methylation-specific polymerase chain reaction (PCR), subtelomeric deletions by MLPA or FISH and Rett syndrome by sequencing the MECP2 gene.
Blood samples were also obtained from 400 healthy blood donors of the same ethnic background as control individuals from the general population.
Screening of mutations in the TBX1 and CRKL genes
We screened for mutations in all exons of the three alternative spliced forms of the gene, TBX1A, TBX1B and TBX1C. We used direct sequencing for mutation scanning. For this purpose, we designed primers that amplify exons and flanking intronic sequences. These primers were designed using the Oligos 9.6 software that can be found at www.biocenter.helsinki.fi/bi/DNA/links.htm). Primer sequences are available on request.
We developed a time- and cost-efficient strategy for the screening of the −39 C → T mutation. This strategy is based on the amplification refractory mutation system (ARMS) technique that uses two reactions, one with primers specific for the normal DNA sequence that cannot amplify mutant DNA and the other with primers specific for the mutant DNA that cannot amplify normal DNA.17 Both reactions contain the same common PCR primer and also a pair of internal control primers that co-amplify a different region of the genome. For this purpose, we used as the common reverse primer 5′-AGTGTAGACACGGCCTGGAG, the wild-type-specific primer: 5′-GCCCACCAGGGCTCAGGGTCCTCCGA**AC and the mutant-specific primer 5′-GCCCACCAGGGCTCAGGGTCCTCCGAAT** (mismatches to the normal sequence shown in bold). As internal control, we used primers that amplify a fragment of exon 9C of TBX1. PCR conditions were the following: 100–150 ng of DNA, 0,2 mM deoxyribonucleotide triphosphates, 20 pmol of each primer, 2 mM of MgCl2, 10% of dimethylsulphoxide and 1 U of AmpliTaq Gold (Applied Biosystems). Cycling conditions were 3′ 95°C, 35 cycles 30′′ 94°C, 1′ 63°C, 45′′ 72°C. PCR products were run on a 2% agarose gel.
In vitro translation and luciferase assay
In vitro translation experiments were performed using the TNT quick-coupled transcription/translation system from Promega, which uses rabbit reticulocyte lysates. The wild-type and mutant TBX1 5′UT regions (129 bp encompassing exon 1 and 2 up to the ATG initiation site with and without the −39 C → T change) were cloned into the _Hind_III–_Not_I sites of the TNT luciferase-T7 plasmid provided with the kit. These constructions resulted in the TBX1 5′UTR downstream from the T7 promoter and immediately upstream of the luciferase initiation site. Transcription and translation experiments were performed following manufacturer instructions. Six independent in vitro transcription and translation experiments for the control plasmid (the luciferase gene without the TBX1 5′UTR) and the wild-type and mutant 5′UTR constructs were carried out. An assay without the plasmid was used as negative control. Results are expressed as relative light units (RLUs).
Results
Mutation screening of the TBX1 gene in patients with 22q11.2-deletion syndrome features
A mutational screen for TBX1 mutations was performed on DNA samples of 38 patients who had congenital heart defect or other clinical features compatible with the 22q11.2-deletion syndrome but with no deletion.
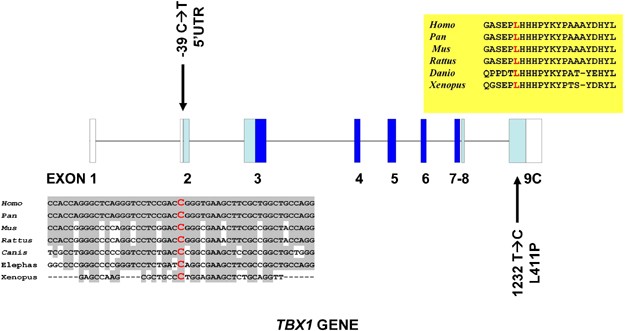
As a result, we found eight previously described common polymorphisms and two rare variants, one of them, which had not been described previously. One of the rare variants is a novel mutation that was localised in exon 9C (1232T>C; L411P) in heterozygosis (Figure 1). The patient is a male child born from a non-consanguineous marriage with a heart outflow abnormality (Tetralogy of Fallot). He has no dysmorphic features, shows a slight T-cell immunodeficiency and a moderate delay in the acquisition of language skills (group 3). The parents were found to be non-carriers and thus it is a de novo mutation. The 1232T>C nucleotide change is nine nucleotides downstream of a previously described frameshift mutation (1223delC) that eliminates a nuclear localisation signal and a transactivation domain essential for the TBX1 function.18 The L411P change is not situated within the identified nuclear localisation signal but affects a residue conserved in all vertebrate species and that consequently seems to be essential for the correct TBX1 function.
Figure 1
Exon–intron structure of the TBX1 gene (transcript C) with the two mutations described in this paper in black. The _trans_-species conservation of the cytosine mutated to a thymine in the 5′UTR is shown in a grey box and that of the leucine residue mutated to proline is shown in a yellow box. Dark blue squares indicate exons encoding the T-box domain, light blue squares exons coding for other TBX1 domains and white squares non-coding regions of the gene.
The other rare variant that was found was a −39 C → T nucleotide change in exon 2 within the 5′UTR (Figure 1). This change was found in two brothers who have Tetralogy of Fallot, one of them suffering from repeated infections and immunodeficiency (group 1 and 3 patients). The −39 C → T mutation within the 5′UTR region had been previously described as a rare variant.15 The authors found this change in a patient with a del22q11.2 syndrome phenotype (absent thymus and a complex conotruncal heart defect) who also carried a 15 bp deletion in exon 9C. Although the −39 C → T change could not be detected in 101 control individuals, it was classified as a rare variant because a healthy brother was also the carrier of both the familial mutations and because the healthy parents carried each one of these mutations. In a similar manner, in the case presented here, we found that the mother of the two children with Tetralogy of Fallot manifests no heart problems or dysmorphic features although she is a carrier. In an attempt to extend our observations, we screened 400 healthy control individuals for the −39 C → T change and found no carriers. This screening was performed using an ARMS methodology. For this purpose, we designed primers that amplify only the −39 C → T mutated allele and primers that amplify only the normal allele. Taking together our data and data reported by Gong et al,15 a total of 2/143 cases with phenotypes encompassing at least one of the del22q11.2 syndrome features have been found to carry the mutation as opposed to 0/501 control individuals (P<0.049; Fischer exact test).
Functional analysis of the −39 C → T mutation
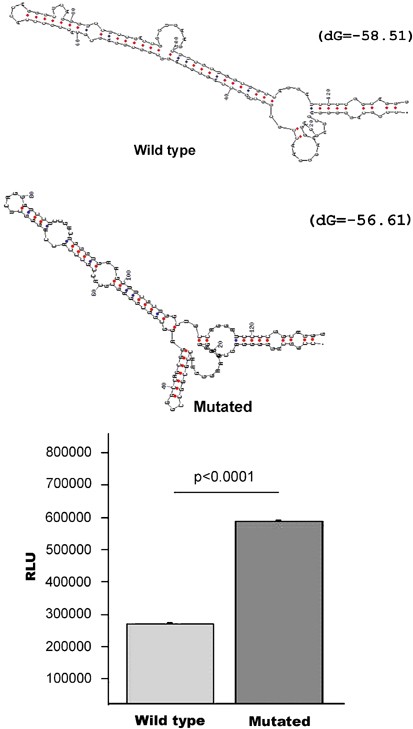
The −39 C → T change affects a cytosine that shows a remarkable _trans_-species conservation for a non-translated base. Such conservation indicates that this 5′UTR cytosine may be critical for the binding of a conserved protein involved in transcription, translation or some other process. Alternatively, this change may affect the secondary structure of the mRNA. A stable secondary structure of the 5′UTR has been shown to inhibit ribosome scanning and initiation of translation. Furthermore, these structures are often found in genes coding for transcription factors, growth factors and proto-oncogenes and deregulation of translation of these RNAs can be associated with disease.19 The 129 bp 5′UTR of the TBX1 gene is GC rich (76% GC) and is predicted to fold into a stable secondary structure by computer modelling using the mfold package (Δ_G_=−58.51) (Figure 2).20 The same computer modelling shows that the presence of a T instead of a C at position −39 decreases the stability of the secondary structure (Δ_G_=−56.61) and results in an optimal folding that is different from that of the wild type.
Figure 2
(a). Computer (mfold) predictions of optimal mRNA structures for the wild-type and mutated (−39 C → T) 5′UTR of the TBX1 gene. (b). Experimental results of in vitro translation experiments performed as described in the text, showing that the mutated 5′UTR results in increased luciferase production (double) (RLU).
To test if the detected −39 C → T 5′UT mutation could affect translation efficiency, we performed in vitro translation experiments. These experiments showed that the insertion of the wild-type 5′UT of TBX1 immediately upstream of the luciferase initiation site greatly decreases luciferase activity (441 × 106 RLU in the control plasmid versus 247 × 105 RLU for the wild-type 5′UT plasmid). However, when we tested the −39 C → T change we observed an increase of luciferase protein levels significantly above the wild-type 5′UT construct (Figure 2) (wild-type 247 × 105±2059 RLU; mutated 527 × 105±3638 RLU; P<0.0001). These results suggest that the mutation described above can potentially increase the production of TBX1 protein at least twofold. As the wild-type and mutant mRNAs were transcribed from the same T7 promoter, the observed luciferase activity increase in the mutant 5′UT is most probably because of a post-transcriptional effect. Furthermore, the reduction in luciferase activity resulting from the insertion of the TBX1 5′UTR is in agreement with reports that a free energy of −50 kcal/mole is sufficient to block ribosome scanning.21 Altogether, these observations suggest that TBX1 protein synthesis is tightly regulated through post-transcriptional mechanisms and that the detected single base change disrupts in some way such regulation.
Exon2 mutation screening of the TBX1 gene in patients referred for fragile-X testing
Patients with 22q11.2. duplication have been described to present mental retardation as one of the phenotypic manifestations.7 Furthermore, recently patients referred for fragile-X determination, because of cognitive and behavioural problems, were found to be carriers of duplications in the 22q11.2 region (2/275 patients).7 Because we found the 5′UT −39 C → T mutation to increase twofold translation efficiency, which could be considered as functionally equivalent to a duplication of the TBX1 gene, we screened a group of patients who had been previously found negative for the FMR1 fragile-X expansion and referred because of cognitive or behavioural disorders. This screen detected 3/200 carriers (P<0.022). The first patient in this group is a 7-year-old boy who shows severe mental and motor retardation, developmental delays (language), hypotonia limbs, obesity, pontocerebellar atrophy, multifocal refractory epilepsy (family background) and dysmorphic features (brachycephaly, micrognathia, hypotelorism, upslanting palpebral fissures, epicanthus, double palpebral folds, small low-set ears). The second patient is an 8-year-old girl with mental and motor retardation, developmental delays, polymicrogyria, asymmetry of lower limbs and dysmorphic features (hypotelorism, upslanting palpebral fissures, down-turned corners of the mouth, broad and depressed nasal root). Both patients suffered frequent infections. The third patient, who is a 15-year-old boy, shows the mildest phenotype of the three and was found to manifest mild mental retardation, aggressive behaviour, advanced puberty, cryptorchidism and anomalies in the hypophysis. The mothers of the first two patients are apparently healthy carriers of the mutation, whereas the parents of the third patient were not available. Thus, we have found that three of 200 patients with cognitive delays carry the −39 C → T change as opposed to 0 of 400 control individuals (501 if we take into account the study by Gong et al15). This carrier frequency is similar to that found in the study by Yobb et al7 for 22q11.2 duplication carriers in fragile X syndrome-testing patients.
Discussion
In the study presented here, we have found two changes in the TBX1 gene that associate with several features of the 22q11. 2 deletion and duplication syndromes. The first mutation, L411P, not previously described and result of a de novo event, was found in a patient with no dysmorphic features, an outflow tract heart defect, T-cell immunodeficiency and mild delay. This mutation affects a conserved amino acid within a putative transactivation domain and is likely to affect the protein's functionality. The second change that we have found is more atypical because it is within the 5′UTR and displays a variable penetrance within the same family. However, we show that it affects translation efficiency in vitro and thus it is likely to have the same effect in vivo altering the TBX1 dosage by increasing it in key steps of development. The −39 C → T 5′UT change in the TBX1 gene can be considered in this sense a functional equivalent to 22q11.2 duplications in the measure that it increases TBX1 protein dosage, an expected outcome for the duplication. The presence of phenotypically normal carrier parents is similar to observations made for the 22q11.2 deletion and duplication syndromes and other TBX1 mutations.2, 7, 15, 22, 23 For example, in a recent study, a phenotypically normal father carrying a 3 Mb 22q11.2 duplication conceived a foetus with the duplication and a complex congenital heart defect and later conceived a normal child also carrying the duplication.2 In addition, phenotypic manifestations caused by already described mutations of the TBX1 gene that inactivate the protein vary widely among affected members of the same families.12, 13 Thus, incomplete penetrance and variable expressivity seems to be a trait of hypomorphic and hypermorphic TBX1 mutations, as well as of 22q11.2 deletions and duplications and indicates that modifying genetic and/or environmental factors are at play.
Recently, it has been described that the products of the TBX1 and CRKL genes, both mapping within the 3 Mb commonly deleted 22q11.2 region, interact in a dose-dependent manner during development.24 The authors show that compound heterozygous mice for null alleles of the TBX1 and CRKL gene show an increase in penetrance and expressivity of del22q11.2 related traits. Previously, it had already been shown that mice homozygous for null alleles of the CRKL gene genocopy multiple aspects of the del22q11.2 syndrome.25 Thus, CRKL is a good candidate to be a modifying gene that affects the expressivity or penetrance of TBX1 mutations. A plausible hypothesis could be that patients affected with the −39 C → T mutation could have inherited the TBX1 gene mutation from one healthy parent and a polymorphism or mutation in the CRKL gene from the other that would exacerbate the clinical manifestations. However, we have sequenced all exons of the CRKL gene in all individuals (healthy parents and affected children) carrying a TBX1 mutation and could not find any mutation or polymorphism (data not shown). Moreover, we searched for significant polymorphisms or mutations in other exons of the TBX1 gene, also without success (data not shown).
There are many other genes that are good candidates to act as modifying genes, for example VEGF, FGF8 or Chordin.26, 27, 28, 29 In this context, −39 C → T mutation carrying individuals should be a good model to search for such modifying factors as we have normal and affected carriers. Moreover, environmental factors may also be important modifiers of the expressivity of TBX1 mutations. For example, TBX1 is inhibited by retinoic acid30, 31 and thus low levels of retinoic acid during development could exacerbate the effects of TBX1 mutations that increase protein levels. Therefore, any environmental factor that interferes with vitamin A metabolism (e.g., alcohol intake) could also potentially affect TBX1 expression during development.32 It is tempting to propose that some of the above mentioned mechanisms could provide explanations for the observation of the incomplete penetrance of the −39 C → T mutation.
In conclusion, we have found two mutations in the TBX1 gene. One of them, L411P, is de novo and associates in a patient with some features of the 22q11.2-deletion syndrome. The other change, −39 C → T, increases TBX1 dosage in vitro and seems to constitute a potential risk for cognitive deficits and heart defects. To our knowledge, this change is the first human mutation showing that TBX1 is a candidate to cause mental retardation associated with the 22q11.2-deletion and -duplication syndromes. In addition, we have determined that three out of 200 patients referred for fragile-X-syndrome screening are carriers of such mutation, which is absent in control individuals. These observations are similar to those made for 22q11.2 duplications, which can be found in patients referred for fragile-X testing and with mental retardation as their sole phenotypic feature. It is probably not coincidental that a 22q11.2 duplication possibly increases TBX1 dosage. This observation suggests that it may be reasonable to screen for the −39 C → T mutation among patients with unspecific cognitive deficits and we provide an easy and quick way to do it with the ARMS approach.
References
- Ryan AK, Goodship JA, Wilson DI et al: Spectrum of clinical features associated with interstitial chromosome 22q11 deletions: a European collaborative study. J Med Genet 1997; 34: 798–804.
Article CAS Google Scholar - de La Rochebrochard C, Joly-Helas G, Goldenberg A et al: The intrafamilial variability of the 22q11.2 microduplication encompasses a spectrum from minor cognitive deficits to severe congenital anomalies. Am J Med Genet A 2006; 140A: 1608–1613.
Article CAS Google Scholar - Ensenauer RE, Adeyinka A, Flynn HC et al: Microduplication 22q11.2, an emerging syndrome: clinical, cytogenetic, and molecular analysis of thirteen patients. Am J Hum Genet 2003; 73: 1027–1040.
Article CAS Google Scholar - Hassed SJ, Hopcus-Niccum D, Zhang L, Li S, Mulvihill JJ : A new genomic duplication syndrome complementary to the velocardiofacial (22q11 deletion) syndrome. Clin Genet 2004; 65: 400–404.
Article CAS Google Scholar - Portnoi MF, Lebas F, Gruchy N et al: 22q11.2 duplication syndrome: two new familial cases with some overlapping features with DiGeorge/velocardiofacial syndromes. Am J Med Genet A 2005; 137: 47–51.
Article Google Scholar - Sparkes R, Chernos J Dicke F : Duplication of the 22q11.2 region associated with congenital cardiac disease. Cardiol Young 2005; 15: 229–231.
Article Google Scholar - Yobb TM, Somerville MJ, Willatt L et al: Microduplication and triplication of 22q11.2: a highly variable syndrome. Am J Hum Genet 2005; 76: 865–876.
Article CAS Google Scholar - Jerome LA, Papaioannou VE : DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet 2001; 27: 286–291.
Article CAS Google Scholar - Lindsay EA, Vitelli F, Su H et al: Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature 2001; 410: 97–101.
Article CAS Google Scholar - Merscher S, Funke B, Epstein JA et al: TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell 2001; 104: 619–629.
Article CAS Google Scholar - Liao J, Kochilas L, Nowotschin S et al: Full spectrum of malformations in velo-cardio-facial syndrome/DiGeorge syndrome mouse models by altering Tbx1 dosage. Hum Mol Genet 2004; 13: 1577–1585.
Article CAS Google Scholar - Paylor R, Glaser B, Mupo A et al: Tbx1 haploinsufficiency is linked to behavioral disorders in mice and humans: implications for 22q11 deletion syndrome. Proc Natl Acad Sci USA 2006; 103: 7729–7734.
Article CAS Google Scholar - Yagi H, Furutani Y, Hamada H et al: Role of TBX1 in human del22q11.2 syndrome. Lancet 2003; 362: 1366–1373.
Article CAS Google Scholar - Conti E, Grifone N, Sarkozy A et al: DiGeorge subtypes of nonsyndromic conotruncal defects: evidence against a major role of TBX1 gene. Eur J Hum Genet 2003; 11: 349–351.
Article CAS Google Scholar - Gong W, Gottlieb S, Collins J et al: Mutation analysis of TBX1 in non-deleted patients with features of DGS/VCFS or isolated cardiovascular defects. J Med Genet 2001; 38: E45.
Article CAS Google Scholar - Rauch A, Devriendt K, Koch A et al: Assessment of association between variants and haplotypes of the remaining TBX1 gene and manifestations of congenital heart defects in 22q11.2 deletion patients. J Med Genet 2004; 41: e40.
Article CAS Google Scholar - Newton CR, Graham A, Heptinstall LE et al: Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res 1989; 17: 2503–2516.
Article CAS Google Scholar - Stoller JZ, Epstein JA : Identification of a novel nuclear localization signal in Tbx1 that is deleted in DiGeorge syndrome patients harboring the 1223delC mutation. Hum Mol Genet 2005; 14: 885–892.
Article CAS Google Scholar - Pickering BM, Willis AE : The implications of structured 5' untranslated regions on translation and disease. Semin Cel lDev Biol 2005; 16: 39–47.
Article CAS Google Scholar - Zuker M : Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003; 31: 3406–3415.
Article CAS Google Scholar - Kozak M : Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol Cell Biol 1989; 9: 5134–5142.
Article CAS Google Scholar - Edelmann L, Pandita RK, Spiteri E et al: A common molecular basis for rearrangement disorders on chromosome 22q11. Hum Mol Genet 1999; 8: 1157–1167.
Article CAS Google Scholar - McDonald-McGinn DM, Tonnesen MK, Laufer-Cahana A et al: Phenotype of the 22q11.2 deletion in individuals identified through an affected relative: cast a wide FISHing net!. Genet Med 2001; 3: 23–29.
Article CAS Google Scholar - Guris DL, Duester G, Papaioannou VE, Imamoto A : Dose-dependent interaction of Tbx1 and Crkl and locally aberrant RA signaling in a model of del22q11 syndrome. Dev Cell 2006; 10: 81–92.
Article CAS Google Scholar - Guris DL, Fantes J, Tara D, Druker BJ, Imamoto A : Mice lacking the homologue of the human 22q11.2 gene CRKL phenocopy neurocristopathies of DiGeorge syndrome. Nat Genet 2001; 27: 293–298.
Article CAS Google Scholar - Abu-Issa R, Smyth G, Smoak I, Yamamura K, Meyers EN : Fgf8 is required for pharyngeal arch and cardiovascular development in the mouse. Development 2002; 129: 4613–4625.
CAS PubMed Google Scholar - Bachiller D, Klingensmith J, Shneyder N et al: The role of chordin/Bmp signals in mammalian pharyngeal development and DiGeorge syndrome. Development 2003; 130: 3567–3578.
Article CAS Google Scholar - Frank DU, Fotheringham LK, Brewer JA et al: An Fgf8 mouse mutant phenocopies human 22q11 deletion syndrome. Development 2002; 129: 4591–4603.
CAS PubMed PubMed Central Google Scholar - Stalmans I, Lambrechts D, De Smet F et al: VEGF: a modifier of the del22q11 (DiGeorge) syndrome? Nat Med 2003; 9: 173–182.
Article CAS Google Scholar - Roberts C, Ivins SM, James CT, Scambler PJ : Retinoic acid down-regulates Tbx1 expression in vivo and in vitro. Dev Dyn 2005; 232: 928–938.
Article CAS Google Scholar - Zhang L, Zhong T, Wang Y, Jiang Q, Song H, Gui Y : TBX1, a DiGeorge syndrome candidate gene, is inhibited by retinoic acid. Int J Dev Biol 2006; 50: 55–61.
Article CAS Google Scholar - Wang XD : Alcohol, vitamin A, and cancer. Alcohol 2005; 35: 251–258.
Article CAS Google Scholar
Acknowledgements
This work was supported by grants from and FIS-PI02654 and FIS-C03/05. We thank Joan Fibla for help with the statistical analysis and for control DNA samples.
Author information
Authors and Affiliations
- Section of Genetics, Hospital Universitari Son Dureta, Palma de Mallorca, Balearic Islands, Spain
Laura Torres-Juan, Jordi Rosell, Catalina Vidal-Pou, Marta Bernues, Angeles Perez-Granero, Nancy Govea & Damian Heine-Suñer - Research Unit, Hospital Universitari Son Dureta, Palma de Mallorca, Balearic Islands, Spain
Montse Morla, Daniel Bachiller & Xavier Busquets - Pediatrics Department, Hospital Universitari Son Dureta, Palma de Mallorca, Balearic Islands, Spain
Fernando García-Algas & Maria-Angeles de la Fuente - Obstetrics and Gynaecology Department, Hospital Fundació Son Llatzer, Palma de Mallorca, Balearic Islands, Spain
Miguel Juan & Albert Tubau
Authors
- Laura Torres-Juan
You can also search for this author inPubMed Google Scholar - Jordi Rosell
You can also search for this author inPubMed Google Scholar - Montse Morla
You can also search for this author inPubMed Google Scholar - Catalina Vidal-Pou
You can also search for this author inPubMed Google Scholar - Fernando García-Algas
You can also search for this author inPubMed Google Scholar - Maria-Angeles de la Fuente
You can also search for this author inPubMed Google Scholar - Miguel Juan
You can also search for this author inPubMed Google Scholar - Albert Tubau
You can also search for this author inPubMed Google Scholar - Daniel Bachiller
You can also search for this author inPubMed Google Scholar - Marta Bernues
You can also search for this author inPubMed Google Scholar - Angeles Perez-Granero
You can also search for this author inPubMed Google Scholar - Nancy Govea
You can also search for this author inPubMed Google Scholar - Xavier Busquets
You can also search for this author inPubMed Google Scholar - Damian Heine-Suñer
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toDamian Heine-Suñer.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary information
Rights and permissions
About this article
Cite this article
Torres-Juan, L., Rosell, J., Morla, M. et al. Mutations in TBX1 genocopy the 22q11.2 deletion and duplication syndromes: a new susceptibility factor for mental retardation.Eur J Hum Genet 15, 658–663 (2007). https://doi.org/10.1038/sj.ejhg.5201819
- Received: 02 November 2006
- Revised: 14 February 2007
- Accepted: 14 February 2007
- Published: 21 March 2007
- Issue Date: June 2007
- DOI: https://doi.org/10.1038/sj.ejhg.5201819