Polymorphism of XRCC1 predicts overall survival of gastric cancer patients receiving oxaliplatin-based chemotherapy in Chinese population (original) (raw)
Introduction
Gastric cancer is the second leading cause of cancer death worldwide, with the highest incidence in China, Japan and Eastern European countries.1 Postoperative chemoradiotherapy has become the standard of care in the treatment of gastric cancer. Fluoropyrimidines, platinums, taxanes and their combinations were shown to be effective in the treatment of gastric cancer. However, the response rates of these drugs or their combinations were less than 50%.2, 3, 4
Individual chemotherapy based on pharmacogenetics and pharmacogenomics has shown a potentially predictive role to achieve superior outcome in retrospective and perspective studies in lung cancer.5, 6 However, little is known in gastric cancer pharmacogenetics. Cisplatin or oxaliplatin is commonly used with 5-fluorouracil (5-FU) as chemotherapy doublets in the treatment of advanced gastric cancer. Genetic polymorphisms in genes involved in DNA repair, drug metabolism and detoxification pathways may influence the activity of these two chemotherapeutic agents, contributing to the individual variability of drug response and toxicity.7
Some researches have demonstrated that suboptimal DNA repair within the tumor may actually lead to the decreased removal of platinum–DNA adducts and, therefore, increased clinical response and may be a prognostic factor for improved survival to platinum chemotherapy.8 Members of the nucleotide excision repair (NER) and base excision repair (BER) pathway are thought to be key genes in DNA repair. Xeroderma pigmentosum group D (XPD) and X-ray repair cross-complementing group 1 (XRCC1), important players in NER and BER pathways respectively, showed potential influence on platinum sensitivity of tumor cells.9 Their functional single nucleotide polymorphisms (SNPs), for example, the XPD Lys751Gln and XRCC1 Arg399Gln, were considered to be predictive factors for patients receiving platinum-based chemotherapy.8
Increasing evidence has suggested an important role for drug-metabolizing enzymes in determining interindividual variations in therapeutic response. Resistance to platinum agents may also depend on altered detoxification pathways. It has been suggested that genetic polymorphisms in certain glutathione _S_-transferase (GST) genes reduce the effectiveness of detoxifying cytotoxins generated by chemotherapeutic agents.10 Glutathione _S_-transferase P1 (GSTP1) plays an important role in detoxification of a variety of chemotherapeutics, including platinum. An A/G SNP located within the substrate-binding domain of GSTP1, which results in valine replacing isoleucine, diminishes GSTP1 enzyme activity.11
As SNP displays ethnic variations, to clarify the impact of polymorphisms in these three genes involved in the detoxification and also DNA repair pathway, we investigated the association between these polymorphisms and overall survival in Chinese gastric cancer patients treated with 5-FU/oxaliplatin combination chemotherapy.
Patients and methods
Patients
Patients included in this study had gastric cancer and were treated with 5-FU plus oxaliplatin as first-line chemotherapy. All patients were administered a modified FOLFOX regimen that consisted of a 2-week cycle of oxaliplatin (85 mg/m2), combined with bolus 5-FU (300 mg/m2), leukovorin (200 mg/m2) and continuous infusion of 5-FU (600 mg/m2).
Overall survival was the end point in the present study. Survival time was calculated from the date of diagnosis to the date of last follow-up or death from any cause. All patients agreed to perform genotype analyses in this study.
Genotyping
Blood sample was collected before chemotherapy, and genomic DNA was extracted from 200 _μ_l whole blood using the QiaAmp kit (Qiagen, Valencia, CA, USA). SNPs in XPD Lys751Gln, XRCC1 Arg399Gln and GSTP1 Ile105Val were assessed by 5′ nuclease allelic discrimination assay (TaqMan) using a fluorescent temperature cycler (Mx3000P Real Time PCR System, Stratagene). Briefly, the 20 _μ_l PCR mixture contained 50 ng of DNA, 1 × absolute QPCR Mix (ABgene, Surrey, UK), 900 nM of primers and 250 nM of probes. The PCR conditions were 50°C for 2 min and 95°C for 15 min, followed by 45 cycles at 95°C for 15 s and 60°C for 1 min. Sequences of primers and probes are available on request. For each SNP, a minimum of 20 randomly selected DNA samples were genotyped at least twice to confirm the results.
Statistical analysis
Demographic and clinical information was compared across genotypes using Fisher's exact test and one-way analysis of variance, where appropriate. The Kaplan–Meier method was adopted to estimate survival curves, and the log-rank test was used to compare patients' survival time between genotype groups. Cox proportional hazard model was used to obtain the _P_-value for genotype corrected by the other clinical parameters, such as ECOG PS (Eastern Cooperative Oncology Group performance status), tumor stage and grade. All _P_-values were two-sided. Statistical significance was defined as P<0.05. Therefore, the significant level with Bonferroni correction was 0.01 (0.05/5). All analyses were performed with the SPSS Version 13.0 software (SPSS Inc., Chicago, IL, USA) and statistical software R (the R project for statistical computing; http://www.r-project.org/).
Results
Patients
A total of 62 gastric adenocarcinoma patients, definitely diagnosed by histology, consisting of 45 men and 17 women with a median age of 55 years, were involved in this study. Of all the patients, 27.4% had stage IIIA, 24.2% had stage IIIB and 48.4% had stage IV disease at the time of diagnosis. ECOG PS was 0–1 in 52 patients, 2 in six patients and 3 in four patients. Detailed demographic and disease characteristics are shown in Table 1. The median survival time (MST) was 322 days (range: 56–2058 days). For patients with ECOG PS≤2, overall survival time ranged from 92 to 2058 days, with an MST of 325 days.
Table 1 Demographic and disease characteristics in gastric cancer patients
Patients' characteristics and their outcomes were unknown to investigators performing genetic analysis. The results of genotyping were disclosed to clinical investigators after data analysis. The study was approved by the ethical committee of Medical School of Nanjing University.
Genotype frequencies of polymorphisms of XPD, XRCC1 and GSTP1
The genotyping of _XPD_-751, _GSTP1_-105 and _XRCC1_-399 were available for all the patients. Wild genotype (A/A) of XPD Lys751Gln was observed in 56 (93.3%) and heterozygous variant (A/C) was present in 6 (6.7%) of 62 patients. Wild genotype (G/G) of XRCC1 Arg399Gln was observed in 20 (32.3%) and heterozygous variant (G/A) in 33 (53.2%), whereas homozygous variant (A/A) was present in 9 (14.5%) of all 62 patients. Wild type (A/A) of GSTP1 Ile105Val was present in 43 (69.4%) and heterozygous variant (A/G) was observed in 19 (30.6%) of all 62 patients. Homozygous variants of codon 751 in the XPD gene and codon 105 in the GSTP1 gene were not observed. Genotype frequencies for XPD, XRCC1 and GSTP1 polymorphisms were found to be in Hardy–Weinberg equilibrium. No significant associations were found between any of these polymorphisms and age, sex, ECOG status, tumor initial stage and grade.
Association between the polymorphisms and overall survival
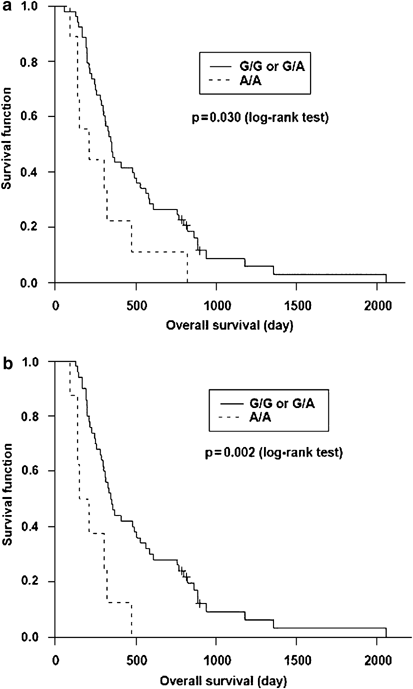
Polymorphisms of XPD and GSTP1 did not show statistically significant survival difference between the homozygous and heterozygous genotypes. Patients with at least one _XRCC1_-399 G allele genotype demonstrated significantly superior MST (337 days and 370 days for G/G and G/A genotypes, respectively) compared to only 212 days for patients with A/A genotype (_P_=0.03). However, the significance was not detected by Cox regression (_P_=0.054). We performed focused analysis on 58 patients with ECOG PS≤2, and more obvious significance was demonstrated (337, 359 and 183 days for G/G, G/A and A/A genotypes respectively; _P_=0.002) (Table 2). Moreover, significance was also demonstrated after Cox regression (_P_=0.0037). Survival curve is shown in Figure 1.
Table 2 Polymorphism of XPD, GSTP1 and XRCC1 and survival in gastric cancer patients
Figure 1
Kaplan–Meier estimates of overall survival by _XRCC1-_399 genotypes: (a) all patients; (b) patients with ECOG PS ≤2. ECOG PS, Eastern Cooperative Oncology Group performance status; XRCC1, X-ray repair cross-complementing group 1.
Discussion
In this study, we assessed three common polymorphisms of the XPD, XRCC1 and GSTP1 genes and determined their association to overall survival to 5-FU/oxaliplatin chemotherapy in gastric cancer patients. We have shown that a G–A transition of the XRCC1 gene at codon 399 was significantly associated with overall survival. Compelling survival differences in this study were apparent in individuals with ECOG PS≤2.
It is still unclear how the change of amino acid at codon 399 of the XRCC1 gene polymorphism influences clinical outcome to oxaliplatin-based chemotherapy in gastric cancer patients. One possible explanation is the enhancement of DNA repair capacity. XRCC1 is thought to be involved in DNA single-strand break repair,12 and it also plays an important role in the BER pathway.13 An SNP in codon 399 was considered to be related to the DNA repair and was likely to exhibit an effect on the protein function.14 Several studies have reported the association of _XRCC1_-399 with the risk in non-small-cell lung cancer (NSCLC),15 colorectal cancer,16 gastric cancer17 and prostate cancer.18 Additionally, reports in colorectal cancer patients show an improved survival for patients with _XRCC1_-399 G allele receiving 5-FU/oxaplatin chemotherapy.19 Similar results were demonstrated in lung cancer,20 breast cancer21 and esophageal cancer.22 Only one recent study has been published, supporting the pharmacogenetic role of _XRCC1_-399 polymorphism in gastric cancer patients.23 The results we report here are in agreement with the current understanding of XRCC1 involvement in platinum compound-based chemotherapy. Our findings support the hypothesis that the outcome of gastric cancer patients receiving platinum compounds may, in part, be due to the polymorphism of DNA repair gene.
Studies have shown the linkage between the _XPD_-751 and _XPD_-312 polymorphisms, two common SNPs of XPD gene.24 Therefore, in the present study, we assessed the SNP at codon 751 and did not find a significant association between _XPD_-751 polymorphism and clinical outcome in gastric cancer patients, supporting previous studies in NSCLC.25, 26 In contrast, Stoehlmacher et al27 reported that patients with C/C genotype showed a notably increased risk of dying, whereas patients with A/C genotype showed an intermediate relative risk. Intriguingly, Dr Rosell and his colleagues reported no significant association between _XPD_-751 polymorphism and overall survival in NSCLC patients in 2004,26 whereas in the recently larger sample-involved study, they showed a significant higher risk of death for patients with C/C genotype.20 However, no C/C genotype was detected in our present study. These results suggest that the polymorphisms might differ according to racial background, which was presumed to occur in previous studies in the Chinese population.28 It was also suggested that larger studies should be carried out to validate the relationship between _XPD_-751 polymorphism and clinical outcome of gastric patients treated with platinum compound-based chemotherapy.
We also found no significant association between _GSTP1_-105 polymorphism and overall survival in gastric cancer patients in Chinese population. It was in agreement with the previous studies of colorectal and breast cancer.27, 29 However, Goekkurt30 and Stoehlmacher31 showed that _GSTP1_-105 G/G genotype predicted superior survival in gastric cancer patients and colorectal cancer patients, respectively. In this study, _GSTP1_-105 polymorphism did not play a significant role in the prediction of clinical outcome, perhaps due to the rare number of patients who had the G/G genotype, indicating that ethnic background eliminated race-specific variation in the distribution of genotypes in GSTP1. Furthermore, studies have shown that GSTP1 is involved in the detoxification of platinum compounds, especially cisplatin, whereas no information is available on the possible impact of GSTP1 on the oxaliplatin pathway. Some differences seem to exist between cisplatin and oxaliplatin detoxification/DNA repair pathways.32 Additionally, there were some controversial reports about the impact of GSTP1 on 5-FU metabolism.33, 34 Therefore, more large-scale researches should be carried out to demonstrate the contribution of GSTP1 in 5-FU/oxaliplatin chemotherapy.
Until now, several studies have focused on the mRNA expressions of DNA repair genes as indicators of chemoresistance to platinum agents. The main obstacle to testing these mRNA expressions is the scarcity of tumor tissue available for RNA isolation and quantitative PCR for formalin-fixed paraffin-embedded tissue. In contrast, it is much easier to obtain DNA isolated from peripheral blood lymphocytes for SNPs analysis. The results we showed here suggest that polymorphism of XRCC1 gene would well be useful as a surrogate marker of clinical outcome in gastric patients with ECOG PS≤2 treated with platinum-based chemotherapy. Further, prospective studies incorporating larger numbers of patients and meta-analysis will be necessary to validate its predicted value.
References
- Parkin DM, Bray F, Ferlay J, Pisani P : Global cancer statistics, 2002. CA Cancer J Clin 2005; 55: 74–108.
Article Google Scholar - Sadighi S, Mohagheghi MA, Montazeri A, Sadighi Z : Quality of life in patients with advanced gastric cancer: a randomized trial comparing docetaxel, cisplatin, 5-FU (TCF) with epirubicin, cisplatin, 5-FU (ECF). BMC Cancer 2006; 6: 274.
Article Google Scholar - Sumpter K, Harper-Wynne C, Cunningham D et al: Report of two protocol planned interim analyses in a randomised multicentre phase III study comparing capecitabine with fluorouracil and oxaliplatin with cisplatin in patients with advanced oesophagogastric cancer receiving ECF. Br J Cancer 2005; 92: 1976–1983.
Article CAS Google Scholar - Van Cutsem E, Moiseyenko VM, Tjulandin S et al: Phase III study of docetaxel and cisplatin plus fluorouracil compared with cisplatin and fluorouracil as first-line therapy for advanced gastric cancer: a report of the V325 Study Group. J Clin Oncol 2006; 24: 4991–4997.
Article CAS Google Scholar - Rosell R, Cecere F, Santarpia M, Reguart N, Taron M : Predicting the outcome of chemotherapy for lung cancer. Curr Opin Pharmacol 2006; 6: 323–331.
Article CAS Google Scholar - Olaussen KA, Dunant A, Fouret P et al: DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med 2006; 355: 983–991.
Article CAS Google Scholar - Marsh S, McLeod HL : Cancer pharmacogenetics. Br J Cancer 2004; 90: 8–11.
Article CAS Google Scholar - Gurubhagavatula S, Liu G, Park S et al: XPD and XRCC1 genetic polymorphisms are prognostic factors in advanced non-small-cell lung cancer patients treated with platinum chemotherapy. J Clin Oncol 2004; 22: 2594–2601.
Article CAS Google Scholar - Weaver DA, Crawford EL, Warner KA, Elkhairi F, Khuder SA, Willey JC : ABCC5, ERCC2, XPA and XRCC1 transcript abundance levels correlate with cisplatin chemoresistance in non-small cell lung cancer cell lines. Mol Cancer 2005; 4: 18.
Article Google Scholar - Hayes JD, Pulford DJ : The glutathione _S_-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol 1995; 30: 445–600.
Article CAS Google Scholar - Watson MA, Stewart RK, Smith GB, Massey TE, Bell DA : Human glutathione _S_-transferase P1 polymorphisms: relationship to lung tissue enzyme activity and population frequency distribution. Carcinogenesis 1998; 19: 275–280.
Article CAS Google Scholar - Brem R, Hall J : XRCC1 is required for DNA single-strand break repair in human cells. Nucleic Acids Res 2005; 33: 2512–2520.
Article CAS Google Scholar - Nazarkina ZK, Khodyreva SN, Marsin S, Lavrik OI, Radicella JP : XRCC1 interactions with base excision repair DNA intermediates. DNA Repair (Amst) 2007; 6: 254–264.
Article CAS Google Scholar - Savas S, Kim DY, Ahmad MF, Shariff M, Ozcelik H : Identifying functional genetic variants in DNA repair pathway using protein conservation analysis. Cancer Epidemiol Biomarkers Prev 2004; 13: 801–807.
CAS PubMed Google Scholar - Kiyohara C, Takayama K, Nakanishi Y : Association of genetic polymorphisms in the base excision repair pathway with lung cancer risk: a meta-analysis. Lung Cancer 2006; 54: 267–283.
Article Google Scholar - Stern MC, Siegmund KD, Conti DV, Corral R, Haile RW : XRCC1, XRCC3, and XPD polymorphisms as modifiers of the effect of smoking and alcohol on colorectal adenoma risk. Cancer Epidemiol Biomarkers Prev 2006; 15: 2384–2390.
Article CAS Google Scholar - Huang WY, Chow WH, Rothman N et al: Selected DNA repair polymorphisms and gastric cancer in Poland. Carcinogenesis 2005; 26: 1354–1359.
Article CAS Google Scholar - Hirata H, Hinoda Y, Tanaka Y et al: Polymorphisms of DNA repair genes are risk factors for prostate cancer. Eur J Cancer 2007; 43: 231–237.
Article CAS Google Scholar - Suh KW, Kim JH, Kim do Y, Kim YB, Lee C, Choi S : Which gene is a dominant predictor of response during FOLFOX chemotherapy for the treatment of metastatic colorectal cancer, the MTHFR or XRCC1 gene? Ann Surg Oncol 2006; 13: 1379–1385.
Article Google Scholar - de las Penas R, Sanchez-Ronco M, Alberola V et al: Spanish Lung Cancer Group. Polymorphisms in DNA repair genes modulate survival in cisplatin/gemcitabine-treated non-small-cell lung cancer patients. Ann Oncol 2006; 17: 668–675.
Article CAS Google Scholar - Bewick MA, Conlon MS, Lafrenie RM : Polymorphisms in XRCC1, XRCC3, and CCND1 and survival after treatment for metastatic breast cancer. J Clin Oncol 2006; 24: 5645–5651.
Article CAS Google Scholar - Wu X, Gu J, Wu TT et al: Genetic variations in radiation and chemotherapy drug action pathways predict clinical outcomes in esophageal cancer. J Clin Oncol 2006; 4: 3789–3798.
Article Google Scholar - Ruzzo A, Graziano F, Kawakami K et al: Pharmacogenetic profiling and clinical outcome of patients with advanced gastric cancer treated with palliative chemotherapy. J Clin Oncol 2006; 24: 1883–1891.
Article CAS Google Scholar - Park DJ, Stoehlmacher J, Zhang W, Tsao-Wei DD, Groshen S, Lenz HJ : A Xeroderma pigmentosum group D gene polymorphism predicts clinical outcome to platinum-based chemotherapy in patients with advanced colorectal cancer. Cancer Res 2001; 61: 8654–8658.
CAS PubMed Google Scholar - Ryu JS, Hong YC, Han HS et al: Association between polymorphisms of ERCC1 and XPD and survival in non-small-cell lung cancer patients treated with cisplatin combination chemotherapy. Lung Cancer 2004; 44: 311–316.
Article Google Scholar - Isla D, Sarries C, Rosell R et al: Single nucleotide polymorphisms and outcome in docetaxel-cisplatin-treated advanced non-small-cell lung cancer. Ann Oncol 2004; 15: 1194–1203.
Article CAS Google Scholar - Stoehlmacher J, Park DJ, Zhang W et al: A multivariate analysis of genomic polymorphisms: prediction of clinical outcome to 5-FU/oxaliplatin combination chemotherapy in refractory colorectal cancer. Br J Cancer 2004; 91: 344–354.
Article CAS Google Scholar - Xing D, Tan W, Wei Q, Lin D : Polymorphisms of the DNA repair gene XPD and risk of lung cancer in a Chinese population. Lung Cancer 2002; 38: 123–129.
Article Google Scholar - Yang G, Shu XO, Ruan ZX et al: Genetic polymorphisms in glutathione-_S_-transferase genes (GSTM1, GSTT1, GSTP1) and survival after chemotherapy for invasive breast carcinoma. Cancer 2005; 103: 52–58.
Article CAS Google Scholar - Goekkurt E, Hoehn S, Wolschke C et al: Polymorphisms of glutathione _S_-transferases (GST) and thymidylate synthase (TS)--novel predictors for response and survival in gastric cancer patients. Br J Cancer 2006; 94: 281–286.
Article CAS Google Scholar - Stoehlmacher J, Park DJ, Zhang W et al: Association between glutathione _S_-transferase P1, T1, and M1 genetic polymorphism and survival of patients with metastatic colorectal cancer. J Natl Cancer Inst 2002; 94: 936–942.
Article CAS Google Scholar - Wang D, Lippard SJ : Cellular processing of platinum anticancer drugs. Nat Rev Drug Discov 2005; 4: 307–320.
Article CAS Google Scholar - Nishiyama M, Yamamoto W, Park JS et al: Low-dose cisplatin and 5-fluorouracil in combination can repress increased gene expression of cellular resistance determinants to themselves. Clin Cancer Res 1999; 5: 2620–2628.
CAS PubMed Google Scholar - Zhan M, Liu X : Schedule-dependent reversion of cisplatin resistance by 5-fluorouracil in a cisplatin-resistant human lung adenocarcinoma cell line A549DDP. Chin Med J (Engl) 1999; 112: 336–339.
CAS Google Scholar
Acknowledgements
This work is supported in part by Research of Health Department in Jiangsu Province (H200640), Medical Technology Development Foundation of Nanjing (ZKX05015) and Clinical Medical Center for Hepatobiliary Disease in Jiangsu Province.
Author information
Author notes
- Baorui Liu and Jia Wei: These authors contributed equally to this work.
Authors and Affiliations
- Department of Oncology, Drum Tower Hospital (affiliated to Medical School of Nanjing University) and Clinical Cancer Institute of Nanjing University, Nanjing, China
Baorui Liu, Jia Wei, Zhengyun Zou, Xiaoping Qian, Wei Zhang, Yitao Ding & Lixia Yu - Laboratory for Statistical Analysis, SNP Research Center, RIKEN (The Institute of Physical and Chemical Research), Tokyo, Japan
Takahiro Nakamura - Department of Oncology, Jiangsu Cancer Hospital, Nanjing, China
Jifeng Feng
Authors
- Baorui Liu
- Jia Wei
- Zhengyun Zou
- Xiaoping Qian
- Takahiro Nakamura
- Wei Zhang
- Yitao Ding
- Jifeng Feng
- Lixia Yu
Corresponding author
Correspondence toBaorui Liu.
Rights and permissions
About this article
Cite this article
Liu, B., Wei, J., Zou, Z. et al. Polymorphism of XRCC1 predicts overall survival of gastric cancer patients receiving oxaliplatin-based chemotherapy in Chinese population.Eur J Hum Genet 15, 1049–1053 (2007). https://doi.org/10.1038/sj.ejhg.5201884
- Received: 07 March 2007
- Revised: 30 May 2007
- Accepted: 30 May 2007
- Published: 27 June 2007
- Issue Date: October 2007
- DOI: https://doi.org/10.1038/sj.ejhg.5201884