Low prevalence of APP duplications in Swedish and Finnish patients with early-onset Alzheimer's disease (original) (raw)
Introduction
Genetics is one of the major risk factors for developing Alzheimer's disease (AD). Four genes are known to be involved in AD and mutations in three of these cause autosomal dominant early-onset Alzheimer's disease (EOAD): the amyloid-β precursor protein (APP) on chromosome 21,1 presenilin 1 (PSEN1) on chromosome 142 and presenilin 2 (PSEN2) on chromosome 1.3 In addition, the ɛ4 allele of apolipoprotein E (APOE) on chromosome 19 increases the risk of developing late-onset sporadic AD and decreases the age at onset in a dose-dependent manner.4 Most of these genetic variants are believed to result in increased production of the amyloid-β (A_β_) peptide or a shift in the A_β_40/A_β_42 ratio to increase the amount of the more amyloidogenic peptide A_β_42. Amyloid-β is cleaved from APP by γ_-secretase whose active constituent is one of the presenilin proteins5 and APOE ɛ4 has been associated with increased A_β deposition.6
Variability in the expression of APP can also cause AD. Individuals with Down's syndrome (DS, trisomy 21) usually develop AD pathology in early middle age due to an extra copy of the APP gene.7 Also, two research groups have recently reported that duplications of APP, in the absence of trisomy 21, can cause familial EOAD with cerebral amyloid angiopathy (EOAD/CAA).8, 9
In this study, we screened for APP duplications in affected subjects from Swedish and Finnish families and cases with EOAD in an effort to estimate the frequency of APP duplications in EOAD patients in these populations.
Materials and methods
Samples
The Swedish sample set consisted of 85 patients from the Memory Clinics of Uppsala Academic Hospital and Karolinska University Hospital, Huddinge. After a quality control, eight samples were removed. The remaining 77 samples with age at disease onset ≤65 years (41–65 years, mean±SD=56.9±6.1 years; 57% women) were included in a screen for APP duplications. All patients were clinically diagnosed as possible or probable EOAD according to NINCDS-ADRDA.10 Autopsy was performed on 17 of these patients, 11 of whom had congophilic vessels. Forty-five patients had familial EOAD with at least one known first-degree relative affected by AD, and 32 were cases without known familial inheritance (Table 1). The Swedish sample collection included the two families with the Swedish (K670N/M671L)11 and Arctic (E693G)12 APP mutations.
Table 1 Samples analyzed for APP gene dose
The Finnish sample set consisted of 64 patients from eastern Finland, all examined at the Kuopio University Hospital, with age at disease onset ≤65 years (45–65 years, mean±SD=59.7±5.3; 72% women). All patients fulfilled clinical criteria for probable AD according to NINCDS-ADRDA.10 Thirty patients had familial EOAD where at least two affected first-degree relatives in two generations had been documented.13 The remaining 34 patients had sporadic EOAD (Table 1). All samples had been screened for mutations in APP, PSEN1 and PSEN2 without positive findings.
This study was approved by the Ethics Committees of Uppsala University and Kuopio University Hospital.
Allele quantification
Primers for _APP_1i (intron after exon 1), _APP_2ei (exon/intron border of exon 2), _APP_9ei (exon/intron border of exon 9), _APP_16i (intron after exon 16), glycogenin 2 (GYG2) on chromosome X and deleted in azoospermia 4 (DAZ4) on chromosome Y were designed using the PrimerExpress software (Applied Biosystems, Foster City, CA, USA); sequences are available upon request. Primers from within the APP promoter, _APP_5e (exon 5) and _APP_18i (the intron after exon 18) were used in Sleegers et al.9 Primers for the control genes hemoglobin beta (HBB) on chromosome 11, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) on chromosome 12 and ubiquitin C (UBC) on chromosome 12, have previously been used in Johnson et al,14 West et al15 and in Sleegers et al,9 respectively.
The Swedish samples were screened for gene load by quantitative PCR (qPCR) using the SYBR green chemistry on the MyiQ Single Color Real-Time PCR Detection System (Bio-Rad, Richmond, CA, USA). The samples were amplified in a volume of 25 μl, containing 10 ng of genomic DNA, 2 × Power SYBR Green PCR Master Mix (Applied Biosystems) and 200 nM of each primer, using the universal thermal cycling parameters (Applied Biosystems). Each plate included triplicate DNA samples from EOAD patients, a healthy control as calibrator sample and two DS patients as positive controls. Wells without template were included as negative controls. SD were calculated for the triplicates. If the SD for a triplicate was more than 0.3, the sample was rerun. The number of gene copies was determined relative to HBB and normalized to the calibrator sample using the 2 -ΔΔ C t method (Applied Biosystems).
The Finnish samples were screened for the presence of APP duplications using both qPCR and a quantitative multiplex PCR (qmPCR) assay. For qPCR, 25 ng of genomic DNA was amplified in a volume of 25 μl together with 2 × Power SYBR Green PCR Master Mix (Applied Biosystems) and 600 nM of each primer (_APP_5e or _APP_16i). Each plate included DNA samples from EOAD patients, four DS patients and four healthy controls, all analyzed in duplicates. The gene dose of APP was calculated using the standard curve method (Applied Biosystems), compared to HBB and normalized to the mean of the healthy controls. The qmPCR was performed in a multiplex PCR using _APP_5e, _APP_16i and HBB where all forward primers were labeled with the fluorescent dye FAM. The reaction mix contained 25 ng of DNA, primers for _APP_5e (200 nM), _APP_16i (100 nM) and HBB (200 nM), 10 × buffer, 10 mM dNTP and 2.5 U HotStarTaq polymerase enzyme (QIAGEN, Hilden, Germany). The number of cycles needed to reach the exponential phase was determined experimentally. Subsequently, 1 μl fluorophore-labeled PCR product was mixed with 10 μl HiDi Formamide and 0.75 μl GeneScan 600 LIZ Size Standard (Applied Biosystems). The PCR product was denatured at 95°C for 3 min and cooled on ice for 2 min. Samples were analyzed through capillary electrophoresis using an ABI 3100 (Applied Biosystems) and the results were viewed using GeneMapper v.3.0 (Applied Biosystems). The peak height was used to determine gene dose of _APP_5e and _APP_16i compared to HBB, and all samples were normalized to the mean of the healthy controls.
SNP arrays
To verify a potential duplication of the APP gene, one sample from the Swedish cohort was run on GeneChip Human Mapping 250K Nsp Arrays (Affymetrix, Santa Clara, CA, USA) according to protocol. The Nsp array has 45 single nucleotide polymorphisms (SNPs) within APP. Results were analyzed with the GTYPE software (Affymetrix).
Results
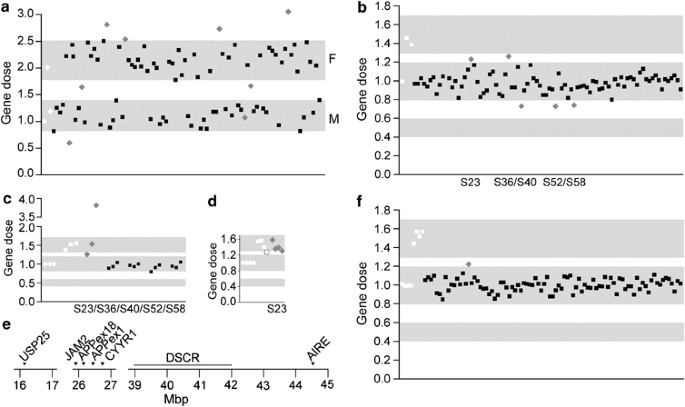
As the qPCR method is dependent on high-quality DNA, the Swedish samples were evaluated by screening with GYG2 on chromosome X. Gene dose data were thereafter related to gender (Figure 1a). Eight samples did not show the expected gene dose on chromosome X and were therefore considered unreliable. They were excluded from further analyses after confirmation of gender with DAZ4 on chromosome Y. The remaining 77 Swedish samples were screened for APP gene dose using qPCR with primers for _APP_16i (Figure 1b) and _APP_1i or _APP_2ei. Initially, five samples with values outside the normal gene dose were detected in the Swedish cohort. Four of the samples were considered normal based on further analysis with additional primers within the APP gene (the promotor, _APP_9ei and _APP_18i) (Figure 1c). One sample still appeared duplicated and the extent of the increased gene dose region was analyzed with two genes flanking APP, JAM2 and CYYR1. Two genes in more distant regions of chromosome 21, USP25 and AIRE, were also analyzed. These genes were compared to HBB (Figure 1d), indicating duplication of one or several regions spanning >28 Mbp on chromosome 21 (Figure 1e). This region also spanned the Down's syndrome critical region (DSCR), the smallest duplicated region of chromosome 21 identified in patients with partial trisomy 21,16 but there were no indications that this patient had DS. To investigate whether this ambiguous duplication could simply be due to a deletion of the control gene, all four genes and _APP_16i were compared to GAPDH and UBC. This did not alter the results. However, the sample demonstrated no duplications of the APP locus when run on a GeneChip 250K SNP array or analyzed using qmPCR.
Figure 1
Gene dose analyses. (a) Quality control of the Swedish samples using GYG2 on chromosome X for correlation to gender, a value of 0.8–1.4 represents men and 1.8–2.5 represents women; (b) Analysis of APP gene dose in the Swedish cohort using primers for intron 16 of APP. Values between 0.4 and 0.6 correspond to a deletion, 0.8–1.2 is normal gene dose and 1.3–1.7 denotes a duplication; (c) follow-up analysis of five Swedish samples; primers for the promotor, intron 9 and intron 18 of APP were used; (d) one Swedish sample in a follow-up analysis of USP25, JAM2, CYYR1 and AIRE; (e) location of the analyzed genes on chromosome 21. Down's syndrome critical region (DSCR) is located in the region of 39–42 Mbp;16 (f) analysis of APP gene dose in the Finnish samples using primers for intron 16 of APP. APP gene dose in the Swedish cohort was determined through qPCR, compared to HBB and normalized to the calibrator sample using the 2 -ΔΔ C t method. All samples were run in triplicates and SD were calculated. If the SD for a triplicate was more than 0.3, the sample was rerun. Number of gene copies in the Finnish cohort was determined through qmPCR, compared to HBB and normalized to the mean of four healthy control samples. White diamonds represent healthy controls, white squares represent Down's syndrome, gray diamonds represent samples outside the specified range and black squares are samples with values within the set gene dose range.
The Finnish samples were screened for APP duplications using _APP_16i and _APP_5e in both qPCR and qmPCR. Seven samples demonstrating increased gene dose in qPCR were re-extracted from peripheral blood lymphocytes and qPCR analysis was repeated. After this, all samples demonstrated normal values. One sample demonstrated a slightly elevated gene dose in qmPCR for both _APP_16i (Figure 1f) and _APP_5e (1.20/1.22), but gene dose was normal (1.02/1.11) in qPCR analysis.
Discussion
This screen for APP duplication included a total of 141 EOAD samples from Sweden and Finland. However, no duplication of the APP locus could be identified. In two recent studies using French8 and Dutch9 samples, the frequencies of APP duplication were calculated to 8 and 2.7% of familial EOAD, respectively. If these frequencies were valid also for the Swedish and Finnish populations, we would have expected to find between two and six cases of APP duplication in our familiar cases. However, the occurrence of APP duplication seems to be more infrequent in the Swedish and Finnish populations.
One sample was concluded to be a false positive finding, as the putative duplication was very large and spanned DSCR, which is not consistent with a non-DS diagnosis.16 There is a hypothetical possibility that this is not an uninterrupted duplication on one chromosome, but rather two independent duplications on separate chromosomes where DSCR is not included. However, this sample did not show any signs of duplication when tested with two other methods, qmPCR and GeneChip SNP arrays.
Neither qPCR nor qmPCR analyses demonstrated evidence of APP gene duplication among the Finnish EOAD patients. Although qmPCR was less variable than qPCR, there were no indications of false negative results using either method, as DS samples consistently demonstrated duplication of chromosome 21. We therefore consider both methods reliable for gene dose screening.
French8 and Dutch9 studies have demonstrated APP duplication ranging from minimally 0.29 up to 6.37 Mb, and containing either only the APP gene9 or up to 12 genes.8 All these duplications were presented with EOAD/CAA. Also, a report on an APP duplication in a Finnish family17 was associated with progressive cognitive decline with coexisting intracerebral hemorrhages, but where the affected family members displayed a cognitive profile somewhat different from that typically seen in AD. Moreover, DS subjects with three copies of chromosome 21, and thereby of APP, usually develop AD although intracerebral hemorrhages have only occasionally been reported.18 These differences in disease presentation could be related to which and how many additional genes are within the duplicated region. Nonetheless, the central role of APP duplications for AD development is emphasized further by a rare case of DS caused by partial trisomy 21 distal to the APP gene. This individual did not show any signs of AD, neither neuropsychological nor neuropathological, at her death at age 78 years.19
In conclusion, we screened a collection of patients from Sweden and Finland with AD and onset at or before 65 years of age, but found no cases of APP duplications. We therefore conclude that duplication of the APP gene is not a common cause of EOAD in Swedish and Finnish populations.
References
- Kang J, Lemaire HG, Unterbeck A et al: The precursor of Alzheimer's disease amyloid A4 protein resembles a cell-surface receptor. Nature 1987; 325: 733–736.
Article CAS Google Scholar - Sherrington R, Rogaev EI, Liang Y et al: Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature 1995; 375: 754–760.
Article CAS Google Scholar - Levy-Lahad E, Wasco W, Poorkaj P et al: Candidate gene for the chromosome 1 familial Alzheimer's disease locus. Science 1995; 269: 973–977.
Article CAS Google Scholar - Corder EH, Saunders AM, Strittmatter WJ et al: Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 1993; 261: 921–923.
Article CAS Google Scholar - Edbauer D, Winkler E, Regula JT, Pesold B, Steiner H, Haass C : Reconstitution of gamma-secretase activity. Nat Cell Biol 2003; 5: 486–488.
Article CAS Google Scholar - Holtzman DM, Bales KR, Tenkova T et al: Apolipoprotein E isoform-dependent amyloid deposition and neuritic degeneration in a mouse model of Alzheimer's disease. Proc Natl Acad Sci USA 2000; 97: 2892–2897.
Article CAS Google Scholar - Olson MI, Shaw CM : Presenile dementia and Alzheimer's disease in mongolism. Brain 1969; 92: 147–156.
Article CAS Google Scholar - Rovelet-Lecrux A, Hannequin D, Raux G et al: APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet 2006; 38: 24–26.
Article CAS Google Scholar - Sleegers K, Brouwers N, Gijselinck I et al: APP duplication is sufficient to cause early onset Alzheimer's dementia with cerebral amyloid angiopathy. Brain 2006; 129: 2977–2983.
Article Google Scholar - McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM : Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology 1984; 34: 939–944.
Article CAS Google Scholar - Mullan M, Crawford F, Axelman K et al: A pathogenic mutation for probable Alzheimer's disease in the APP gene at the N-terminus of beta-amyloid. Nat Genet 1992; 1: 345–347.
Article CAS Google Scholar - Nilsberth C, Westlind-Danielsson A, Eckman CB et al: The ‘Arctic’ APP mutation (E693G) causes Alzheimer's disease by enhanced Abeta protofibril formation. Nat Neurosci 2001; 4: 887–893.
Article CAS Google Scholar - Lehtovirta M, Soininen H, Helisalmi S et al: Clinical and neuropsychological characteristics in familial and sporadic Alzheimer's disease: relation to apolipoprotein E polymorphism. Neurology 1996; 46: 413–419.
Article CAS Google Scholar - Johnson J, Ostojic J, Lannfelt L et al: No evidence for tau duplications in frontal temporal dementia families showing genetic linkage to the tau locus in which tau mutations have not been found. Neurosci Lett 2004; 363: 99–101.
Article CAS Google Scholar - West AB, Kapatos G, O'Farrell C et al: N-myc regulates parkin expression. J Biol Chem 2004; 279: 28896–28902.
Article CAS Google Scholar - Sinet PM, Theophile D, Rahmani Z et al: Mapping of the Down syndrome phenotype on chromosome 21 at the molecular level. Biomed Pharmacother 1994; 48: 247–252.
Article CAS Google Scholar - Rovelet-Lecrux A, Frebourg T, Tuominen H, Majamaa K, Campion D, Remes AM : APP locus duplication in a Finnish family with dementia and intracerebral haemorrhage. J Neurol Neurosurg Psychiatry 2007; 78: 1158–1159.
Article CAS Google Scholar - Cabrejo L, Guyant-Marechal L, Laquerriere A et al: Phenotype associated with APP duplication in five families. Brain 2006; 129: 2966–2976.
Article Google Scholar - Prasher VP, Farrer MJ, Kessling AM et al: Molecular mapping of Alzheimer-type dementia in Down's syndrome. Ann Neurol 1998; 43: 380–383.
Article CAS Google Scholar
Acknowledgements
We express our appreciation to all the families who participated in this study and to all the people involved in the collection of the samples, including Dr Marie-Louise Bondeson at the Department of Genetics and Pathology, Uppsala University, Sweden, for the Swedish Down's syndrome samples. We also thank Professor Christine Van Broeckhoven at Department of Molecular Genetics, University of Antwerp, Belgium, for providing us with primer sequences for the promoter of APP, _APP_5e and _APP_18i. This work was supported by The Swedish Research Council, The Swedish Alzheimer foundation, APOPIS (Contract no. LSHM-CT-2003-503330), Gun and Bertil Stohnes foundation, the Dementia foundation, EVO Grant 5772708 from Kuopio University Hospital, the Marie Curie Early Researcher Program, BiND, University of Kuopio (JV) and by the EU regional funds project no. 86428.
Author information
Author notes
- Elin S Blom, Jayashree Viswanathan, Anna Glaser and Mikko Hiltunen: These authors have contributed equally to this work.
Authors and Affiliations
- Department of Public Health and Caring Sciences, Section of Molecular Geriatrics, Uppsala University, Uppsala, Sweden
Elin S Blom, Lena Kilander, Lars Lannfelt, Martin Ingelsson & Anna Glaser - Department of Neuroscience and Neurology, University of Kuopio, K, u, opio, Finland
Jayashree Viswanathan, Seppo Helisalmi, Hilkka Soininen & Mikko Hiltunen
Authors
- Elin S Blom
- Jayashree Viswanathan
- Lena Kilander
- Seppo Helisalmi
- Hilkka Soininen
- Lars Lannfelt
- Martin Ingelsson
- Anna Glaser
- Mikko Hiltunen
Corresponding author
Correspondence toElin S Blom.
Rights and permissions
About this article
Cite this article
Blom, E., Viswanathan, J., Kilander, L. et al. Low prevalence of APP duplications in Swedish and Finnish patients with early-onset Alzheimer's disease.Eur J Hum Genet 16, 171–175 (2008). https://doi.org/10.1038/sj.ejhg.5201966
- Received: 05 July 2007
- Revised: 10 October 2007
- Accepted: 31 October 2007
- Published: 28 November 2007
- Issue Date: February 2008
- DOI: https://doi.org/10.1038/sj.ejhg.5201966