Escherichia coli in Infants' Intestinal Microflora: Colonization Rate, Strain Turnover, and Virulence Gene Carriage (original) (raw)
Main
Escherichia coli is one of the first bacterial species to colonize the infant's intestines. In the 1970s, E. coli usually appeared in the baby's feces a few days after birth (1, 2), as a sign of its establishment in the intestinal microflora (3, 4). E. coli colonizing the newborn infant may originate in the maternal fecal flora (5), but E. coli strains are also commonly spread at maternity wards via the nursing staff, especially during periods of high bed-occupancy and staff workload (6). We have recently reported that Staphylococcus aureus has become a major colonizer of the infant gut (7), which may be a sign of reduced competition from other microbes. E. coli and other fecal bacteria might be less easily spread today, because of increased hygiene in hospitals and families.
Some E. coli strains persist in the intestinal microflora of an individual for months or years (resident strains), whereas others (transient strains) disappear within a few weeks (8). Resident E. coli strains display certain characteristics that enable them to persist in the intestinal microflora, e.g. the expression of P fimbriae and capacity to adhere to colonic epithelial cells (9–12). P fimbriae are composed of a fimbrial rod with a tip adhesin that exists in three varieties, termed papG classes I, II, and III. These recognize the Galα1→4Galβ disaccharide, with slight differences in binding specificity (13). The class II variety of the papG adhesin is common among E. coli causing pyelonephritis (14), whereas the class III variety is common in cystitis strains from humans (15), dogs (16), and cats (17). Intestinal persistence of E. coli has been linked to the class II variety of the adhesin (10). E. coli strains resident in the human colonic microflora also more commonly possess genes for other virulence factors, such as the iron-chelating compound aerobactin (9, 10), and the capsular types K1 and K5, compared with colonic transient strains (10).
The present study was designed to investigate the E. coli colonization pattern during the first year of life in Swedish infants born in the late 1990s, in relation to delivery mode and lifestyle factors, such as feeding pattern, family size, and pet ownership. Individual E. coli strains were identified, and their possession of genes for adhesins and other virulence factors were related to persistence in the intestinal microflora.
METHODS
Subjects.
Seventy infants (35 girls and 35 boys) born in 1998-1999 at the Sahlgrenska University Hospital, Sweden, were included. They were part of a prospective birth-cohort study aiming to examine the relation between intestinal colonization pattern and allergy development, and 60 of 70 infants had at least one allergic parent. Information was obtained about siblings and pets, and the parents recorded the baby's feeding pattern. The records were checked by a study nurse who interviewed the parents by telephone at 6 and 12 mo. Informed consent was obtained, and the study was approved by the Medical Ethics Committee of Göteborg University.
Nine infants were delivered by cesarean section owing to signs of fetal asphyxia (n = 3), maternal infection (n = 1), preeclampsia (n = 1), myoma (n = 2), pelvic contraction (n = 1), or psychological reasons (n = 1). Most infants roomed in with their mothers at the maternity ward directly after delivery and left the hospital after an average of 2.5 d. Seven infants stayed a few days at the neonatal ward before being transferred to the maternity ward.
All mothers commenced breast-feeding, but five infants were breast-fed for less than 2 mo, and thereafter received commercial formula. Solids were mostly introduced by 4 mo of age, but 77% of the infants received breast milk in addition to solids until at least 6 mo of age. Fruits and vegetables were regularly introduced between 4 and 5 mo of age, porridge and gruel at 4-6 mo of age, and meat at 5-6 mo of age. Fish and egg were rarely introduced before 6 mo of age.
Isolation of E. coli from infants' intestinal microflora.
A sample of the rectal flora was obtained 3 d after delivery using a cotton-tipped swab. The swab was put in COPAN's transportation medium (18) and transported to the laboratory within 20 h. The swab was streaked on a Drigalski agar plate (19), and the inoculate was spread to obtain free-lying colonies.
Fecal samples were obtained at 1, 2, 4, and 8 wk of age and at 6 and 12 mo of age. Feces was collected at home by the parents and put in a plastic bag in which an anaerobic atmosphere was generated (AnaeroGen Compact, Oxoid, Hampshire, U.K.). The samples were kept refrigerated until being transported to the laboratory where they were processed within 24 h after collection. Using this procedure, E. coli counts were not reduced compared with immediate culture as shown in preliminary experiments.
Feces was diluted serially, plated on Drigalski agar (19), and incubated aerobically overnight at 37°C. In each sample, individual colony types differing in size, shape, color. or mucoid appearance were separately enumerated and subcultured for purity, and their species identity was determined using the API20E biotyping system (API Systems SA, La Balme les Grottes, Montalieu-Vercieu, France). Colony types representing E. coli were saved at −70°C in Hogner's freezing medium (20) and analyzed further.
The limit of detection was 330 cfu/g feces (represented logarithmically as 102.52). Subdominant strains could be identified if they differed in morphology and if their population numbers differed less than two log units from the dominant colony type. Colony types less frequent than this were overgrown and missed. One to six colony types were regularly identified in each sample.
Strain typing by RAPD.
Different E. coli strains were identified by RAPD (21). A small amount of bacteria obtained from an overnight culture on tryptic soy agar (TSA) plates was added to 50 μL of HotStarTaq Master Mix, (Qiagen, Spånga, Sweden) containing 6 μM of the primer GTGATCGCAG. The PCR was started with a 95°C 15-min heat activation step for the polymerase, and continued with the following temperature profile: 94°C for 45 s; 30°C for 120 s; 72°C for 60 s for four cycles followed by 94°C for 5 s; 36°C for 30 s; 72°C for 30 s for 26 cycles (the extension step was increased by 1 s for every new cycle). The reaction was terminated at 72°C for 10 min and cooled to 4°C. The PCR products were separated on 8% ready-made Tris-Glycine gels and visualized by silver staining (21).
All E. coli isolates from one child were assayed together, and their PCR products were, when possible, separated on the same gel. Two isolates with identical profiles were considered to belong to the same strain. If two isolates showed minor differences in RAPD patterns, they were run together in a new PCR and thereafter assigned to the same or different strains.
Multiplex PCR for detection of virulence genes in E. coli strains.
Virulence factor genes were identified using three sets of multiplex PCRs, the first identifying the genes for type 1 fimbriae (fimA), P fimbriae (papC), S fimbriae (sfaD/E), and Dr. hemagglutinin (draA); the second the class I, class II, and class III varieties of the P-fimbrial adhesin gene papG; and the third the genes for the capsule K1 (neuB) and K5 (kfiC), aerobactin (iutA), and hemolysin (hlyA) (10). The primer pairs used have been published previously (10, 22–25).
The multiplex PCRs were performed as previously described (10), but with slight modifications. Bacteria from colonies grown on TSA were added to a mixture containing HotStarTaq Master Mix (Qiagen) and 0.45 μM of each primer pair in a final volume of 50 μL. In the third multiplex PCR reaction, the concentration of MgCl2 was increased from 1.5 to 2.0 mM. The PCR program was started with an initial heat activation step for the Taq polymerase at 95°C for 15 min. Thereafter, the PCR was run as described previously (10). PCR products were separated by agarose gel electrophoresis and stained with ethidium bromide.
Statistical methods.
Proportions were compared using Fisher's exact test. Population counts were compared using the Mann-Whitney U test.
RESULTS
Colonization by E. coli.
Forty-two percent of the 70 infants were colonized by E. coli 3 d after birth. After this, further colonization occurred slowly. It took more than 2 wk before half of the infants had E. coli in their stools and by 2 mo only 61% were colonized. One child had not yet acquired any E. coli at 1 y of age, and another eight infants (11%), who had earlier been colonized with E. coli, were again negative by 1 y of age.
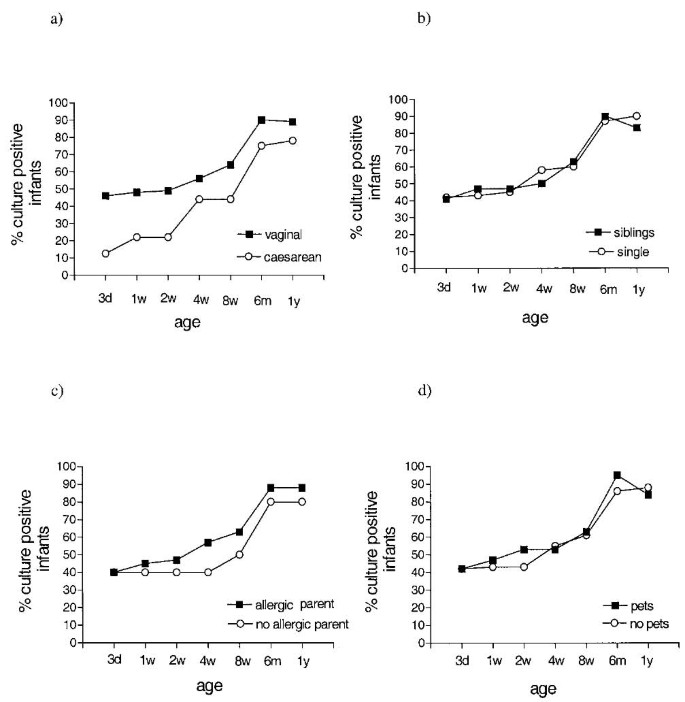
Infants delivered by cesarean section showed delayed acquisition of E. coli compared with vaginally delivered infants (Fig. 1A), although the difference between the groups did not reach statistical significance at any time. There was no difference in colonization rate between infants with or without siblings, with allergic or nonallergic parents, or between infants who grew up in a home with or without pets (Fig. 1, B-D).
Figure 1
Intestinal E. coli colonization rate in infants delivered vaginally (n = 61) or by cesarean section (n = 9; A); infants with (n = 30) or without siblings (n = 40; B); infants with allergic (n = 60) or nonallergic parents (n = 10; C); and infants growing up in a home with (n = 19) or without pets (n = 51; D).
Populations counts of E. coli.
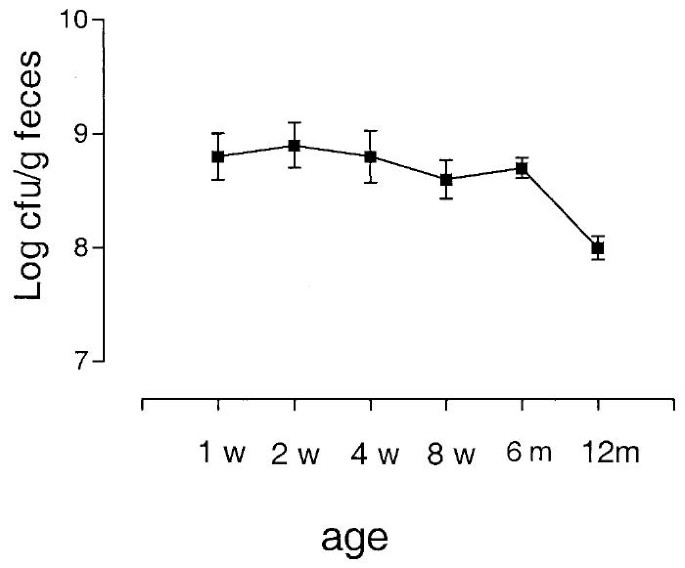
In colonized infants, the population counts of E. coli were high during the first 6 mo of life (Fig. 2). A significant decrease in E. coli population numbers occurred between 6 and 12 mo of age (from 108.7 cfu/g to 108.0 cfu/g, n = 61, 60, p < 0.0001). E. coli population levels did not differ at 6 mo of age between infants receiving or not receiving breast milk at that age (108.7 cfu/g feces versus 108.6 cfu/g, n = 47, 14, p = 0.72).
Figure 2
Total E. coli counts in fecal samples from culture-positive infants. The mean and SD is given for each time point.
Strain turnover.
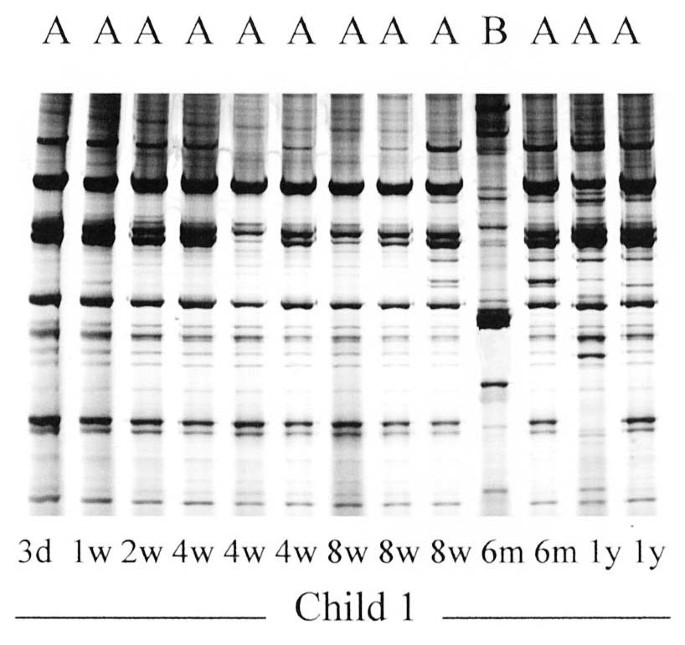
Individual strains of E. coli in an infant were identified by RAPD. Strain identities were not compared among infants. Figure 3 shows the RAPD profiles of the E. coli isolates obtained from one infant. Strain “A” was present on all sampling occasions from d 3 and onward, whereas another strain “B” was only present in the 6-mo sample. Although several colonies were often selected from a sample, because they differed in morphology and were suspected to represent different strains, they often turned out to be identical by RAPD (e.g. the three isolates in the 4-wk sample).
Figure 3
RAPD patterns of 13 intestinal E. coli isolates obtained on different occasions from a single child. Strain “A” was present on all sampling occasions, whereas strain “B” was present in the 6-mo sample only. The three isolates on the right end of the gel were reanalyzed and thereafter assigned to strain “A.”
During the first 6 mo of life, 13% of the infants had no E. coli, 53% carried a single E. coli strain, 19% had two, 9% had three, and 7% had four to six strains. On average, 1.5 strains per child were identified during the first 6 mo.
During the first year of life, 1.4% had no E. coli, 39% carried a single strain, 31% had two strains, 16% had three strains, and 13% had four to six strains. On average, 2.1 different E. coli strains were found per infant during the first year of life. The diversity of the E. coli flora in colonized infants did not increase with age. Thus, at 3 d and at 12 mo of age, each infant positive for E. coli harbored on average 1.2 different strains. The mean number of strains isolated in the first year was equal in children who were delivered by cesarean section (n = 9) or vaginally (n = 61; 2.1 versus 2.1) as well as in children with (n = 30) or without (n = 40) elder siblings (2.2 versus 2.1). Infants growing up in a household with pets (n = 19) had no higher turnover rate of E. coli in their microflora than infants in pet-free households (n = 51; 1.8 versus 2.2; p = 0.53). Children of allergic parents (n = 60) had slightly, but insignificantly, fewer strains than children of nonallergic parents (n = 10; 2.0 versus 2.7; p = 0.42).
Five infants ceased breast-feeding before 2 mo of age, 11 between 2 and 6 mo, and 54 still received breast milk at 6 mo of age. Infants in the first and last groups acquired equally few E. coli strains during the period 2-6 mo (0.4 strains per infant).
Genes for adhesins and other virulence factors in intestinal E. coli strains.
One isolate of each E. coli strain was analyzed by multiplex PCR for the detection of genes for virulence factors. In addition, several isolates from 17 strains were analyzed to assess the consistence of virulence gene carriage. All isolates of the 17 strains were identical with respect to virulence factor genes.
Among the 149 strains, genes encoding type 1 fimbriae were carried by 71%, genes for P fimbriae by 32%, and for S fimbriae by 26%. Not a single strain carried the genes for Dr hemagglutinin. Among the P-fimbriated strains, none carried the class I adhesin, 45% (21 of 47) carried the class II adhesin only, 34% (16 of 47) carried the class III adhesin only, and 8.5% (4 of 47) carried both the class II and class III adhesins, whereas 13% (6 of 47) lacked any of these adhesin varieties. Genes encoding the K1 capsule were found in 27%, the K5 capsule in 7%, aerobactin in 29%, and hemolysin in 23% of the strains.
Certain combinations of virulence genes tend to occur in combinations. We investigated virulence gene associations in the 149 E. coli strains. The P-fimbrial gene papC was significantly associated with the S-fimbrial gene sfaD/E (p = 0.0021), the hemolysin gene hlyA (p < 0.0001), the aerobactin gene iutA (p = 0.0061), and the gene for the K1 capsule neuB (p = 0.0014). papG class II was associated with hlyA (p = 0.036), iutA (p = 0.0028), and neuB (p = 0.0001), whereas papG class III was associated with sfaD/E (p = 0.0001) and hlyA (p < 0.0001). The S-fimbrial gene sfaD/E was significantly associated with hlyA (p < 0.0001) and neuB (p = 0.019). The aerobactin gene iutA was significantly associated with the K1 gene neuB (p = 0.001).
Strains recovered from 0- to 2-mo-old infants (n = 71) were compared with those recovered from 6- to 12-mo-old infants (n = 78). The former slightly more often possessed genes for K1 (34% versus 21%) and aerobactin (35% versus 23%) than the latter, but the differences were not significant (p = 0.095 and p = 0.11, respectively). The other virulence factor genes were equally common in early and late strains.
Strains first appearing in the microflora during exclusive breast-feeding (n = 66) were compared with strains acquired after termination of breast-feeding (n = 48). Genes for the K1 capsule were significantly more common in the strains acquired by exclusively breast-fed compared with weaned infants (32% versus 15%, p = 0.047). Carriage of genes for other virulence factors was similar in the two groups.
E. coli strains recovered from the five children who had a cat in the family carried papG allele III (42% versus 11%, p = 0.011) and the gene for hemolysin (59% versus 18%, p = 0.021) significantly more often than strains from children whose families had no pets. Strains isolated from children in households with a dog did not differ significantly from strains isolated from infants without pets.
Virulence factor genes in resident and transient E. coli strains.
Forty-seven of the 70 infants harbored at least one E. coli strain that persisted in the microflora for at least 3 wk. These strains were classified as resident. Strains persisting for shorter periods were defined as transient, but strains appearing only in the 2-, 6-, or 12-mo sample were neither defined as resident nor transient, because several months passed between these sampling occasions. A total of 58 E. coli strains were classified as resident and 19 as transient. The mean time of persistence for the resident strains was 30 wk. Because half of the strains were still present at 12 mo of age, this is an underestimation of the persistence time.
The carriage rate of different virulence factor genes in resident and transient strains is shown in Table 1. Genes for type 1 fimbriae, P fimbriae, and hemolysin were all significantly more common in resident than in transient strains. Furthermore, a combination of genes for P fimbriae and type 1 fimbriae was 7 times more prevalent among resident than among transient strains (p = 0.0088) and was, thus, more strongly associated with persistence than either P or type 1 fimbriae alone.
Table 1 Carriage of genes encoding different virulence factors in resident and transient intestinal E. coli strains*
Several other combinations of virulence factor genes were associated with persistence, but in these cases the combination was not more strongly associated with persistence than one of the genes alone. Combinations of virulence factors that were more common in resident than transient strains included S fimbriae and hemolysin (12 of 58 versus 0 of 19, ratio >4.1, p = 0.032), P fimbriae and hemolysin (14 of 58 versus 0 of 19, ratio >4.8, p = 0.016), P and S fimbriae (11 of 58 versus 0 of 19, ratio >3.8, p = 0.056), and type 1 fimbriae and hemolysin (13 of 58 versus 0 of 19, ratio >4.5, p = 0.030). Overall, a combination of four or more virulence factors was detected in 36% of the resident versus 5.3% of the transient strains (ratio, 6.8; p = 0.0088). Conversely, only 5% of the resident strains carried neither of the tested virulence factors, as compared with 37% of the transient strains (ratio, 0.14; p = 0.0015).
DISCUSSION
In the present study, the intestinal E. coli colonization pattern was characterized in 70 healthy Swedish infants born in the late 1990s. E. coli is a bacterium which can only thrive in the intestines of man and other animals (26). It can, thus, be spread only via fecal contamination. Such contamination may occur during a vaginal delivery, and 45% of vaginally delivered, but only 12% of cesarean section delivered, infants were positive for E. coli by 3 d of age. This suggests that approximately one third of infants acquired E. coli from their mother at delivery, which fits well with other studies (27, 28).
In the 1970s, at least 70% of infants born in Western countries acquired E. coli during their first week of life (1, 2). Strains spread in the hospital milieu contributed significantly to colonization (29). In developing countries, almost all infants acquire E. coli in the first week of life (30, 31), including infants delivered by cesarean section (31, 32) In the present study, we noted less than 50% E. coli colonization by 1 wk of age. Spread of E. coli in maternity wards is restricted by rooming-in (33). We believe that rooming-in combined with early discharge from the hospital have reduced early colonization by E. coli.
It took 6 mo before (almost) all infants were colonized by E. coli, which is later than observed in the 1980s (34). This slow acquisition was not caused by the fact that most infants in our study had an allergic parent(s), because colonization occurred equally slowly in those with allergic and nonallergic parents. Siblings or pets in the household also did not influence E. coli colonization rate.
Once colonized, the studied infants displayed a slow turnover rate of individual E. coli strains in the microflora. During the first 6 mo of life, a mean number of 1.5 strains per infant were identified, which contrasts sharply to the 8.5 strains found on average in Pakistani infants (35). One may also compare the 2.1 different E. coli strains found during the first 12 mo in our study with the average 4.2 strains found in Swedish infants followed until 11 or 18 mo of age during the 1980s (36). Differences in sampling schedules and number of selected colonies preclude a direct comparison, but one may conclude that the turnover of individual E. coli strains in Swedish infants is at least as low as it was in the 1980s, and might even have further decreased since then.
We did not observe any greater number of E. coli strains in the microflora of children who shared households with older siblings or pets. We also did not observe any greater turnover of E. coli strains between 2 and 6 mo of age in weaned compared with breast-fed children, indicating that feeds were not a significant source of E. coli strains. In contrast, in a study performed in the 1980s breast-fed infants harbored fewer E. coli strains over time than bottle-fed infants (37).
The normal intestinal microflora is the major drive for the maturation of the intestinal immune system (38). A bacterial strain that colonizes the gut activates the gut immune system only transiently, even if it persists in the microflora. The secretory IgA induced in response to the bacterium coats it and prevents it from translocating across the mucosal barrier and, hence, from interacting further with the immune system (39). A constant turnover of strains in the microflora may, therefore, be needed to keep the immune system activated. In accordance, Pakistani infants, who are constantly exposed to new E. coli strains, have a strong secretory IgA response against a pool of E. coli O antigens already at 2 mo of age, whereas Swedish infants do not reach similar levels until 1-2 y of age (40). The total secretory IgA concentration in saliva also rises much more rapidly in Pakistani than Swedish infants (40).
E. coli population numbers remained high during the first 6 mo of life, but decreased 10-fold between 6 and 12 mo of age. The populations of facultative bacteria diminish as the anaerobic microflora becomes more complex (41, 42). This has been reported to occur already in the first weeks in earlier studies (30, 43). The late decrease in E. coli population numbers seen here could reflect delayed acquisition of a complex anaerobic microflora in the Swedish infants born today. We have noted very high fecal numbers of S. aureus (7) and coagulasenegative staphylococci (unpublished observations) in the infants examined here, which we interpret as a sign of reduced competition from other bacteria in the intestinal microflora.
Virulence genes were identified in the E. coli strains colonizing the infants' intestines. The carriage rates of these genes were similar to those observed in E. coli isolated from the intestinal flora of Swedish schoolgirls in the 1970s (10), but higher than among E. coli isolated from Pakistani infants (9), which confirms that commensal E. coli populations may differ in different geographic areas (35, 44).
Analysis of E. coli virulence gene carriage in relation to lifestyle factors revealed that E. coli acquired during exclusive breast-feeding significantly more often had the gene for K1 than those acquired during formula-feeding. This could be an age effect, as K1 tended to be more common in E. coli strains acquired during the first 2 mo than in strains acquired later, but it could also be the other way around; the tendency of K1 strains to colonize newborn infants could be related to the fact that they are often breast-fed. Previous studies have instead found the reverse: a decreased colonization by E. coli carrying K1 as well as P fimbriae in breast-fed infants (45, 46).
Another finding was that the class III variety of the papG adhesin located on P fimbriae, as well as the gene for hemolysin, were both significantly more common in E. coli in the microflora of children with a cat in the family, compared with E. coli isolated from children growing up in pet-free households. One may speculate that E. coli from a family cat could be transferred to the infant, because E. coli causing urinary tract infection in cats and dogs frequently carry the papG III adhesin type, and these strains arise from the intestinal flora of the animals (16, 17). We saw no effect on E. coli gene carriage by dog-ownership. Possibly the parents allow cats to have closer contact with the infant compared with dogs. Infants growing up with pets have lesser risk of developing allergy (47, 48), which might result from exposure to microbes with protective properties.
Resident and transient strains were compared for virulence gene carriage rates. We have previously shown that E. coli strains persisting in the intestinal microflora more often carry the genes for P fimbriae, K1 or K5 capsule, and aerobactin than transient strains in the same hosts (9, 10). In the present study, P fimbriae and type 1 fimbriae, and especially the combination of these two adhesins, were significantly associated with intestinal persistence. P fimbriae and type 1 fimbriae confer adherence to colonic epithelial cells (49), and type 1 fimbriae bind to the carbohydrate chains of secretory IgA, a major component of breast-milk (50). Interaction with secretory IgA seems to enhance intestinal survival of type 1-fimbriated E. coli, because these are reduced in IgA-deficient individuals (51).
The gene for hemolysin was also much more common among resident than transient strains. Hemolysin, which is toxic to a number of cell types (52), enhances colonization by diarrheogenic E. coli in the pig small intestine (53). Hemolysin might contribute to persistence by attacking enterocytes and releasing nutrients for the bacteria. In this context, it is interesting to note that E. coli in the large intestine appears to use membrane lipids as its main nutrient source (54). The association between hemolysin and persistence could also be indirect, as several different virulence factor genes occur together, sometimes carried on so-called pathogenicity islands in the genome (55–57). Indeed, 36% of the resident strains carried four or more virulence factor genes, compared with 5.3% of the transient strains. Pathogenic E. coli belong to certain phylogenetic subgroups (58). Further studies may determine whether the strains that are successful in colonizing the large intestine belong to the same subgroups, and whether they possess pathogenicity islands. These might rather be termed “fitness islands,” as previously suggested (59), if they first aid in intestinal colonization and then confer pathogenicity to E. coli reaching extraintestinal sites.
CONCLUSIONS
In conclusion, the results of the present study demonstrate that E. coli is acquired late in Swedish infants today and that the turnover rate of individual strains is low, most likely because of a limited circulation of fecal bacteria in the Swedish hospitals and homes. This reduced bacterial exposure might have consequences for the maturation of the immune system and its function. Despite indications of poor competition within the intestinal ecosystem, E. coli strains that colonized for prolonged periods possessed certain traits that were much less common in transient strains. These traits, traditionally regarded as virulence factors, have most likely evolved to increase the fitness of E. coli in the human colon.
Abbreviations
CFU:
colony-forming units
RAPD:
random amplified polymorphic DNA
REFERENCES
- Hewitt JH, Rigby J 1976 Effect of various milk feeds on numbers of Escherichia coli and Bifidobacterium in the stools of new-born infants. J Hyg (Lond) 77: 129–139
Article CAS Google Scholar - Feeney AR, Cooke EM, Shinebaum R 1980 A comparative study of gram-negative aerobic bacilli in the faeces of babies born in hospital and at home. J Hyg (Lond) 84: 91–95
Article CAS Google Scholar - Nelson DP, Mata LJ 1970 Bacterial flora associated with the human gastrointestinal mucosa. Gastroenterology 58: 56–61
CAS PubMed Google Scholar - Moore WEC, Cato EP, Holdeman LV 1978 Some current concepts in intestinal bacteriology. Am J Clin Nutr 31:S33–S42
Article CAS PubMed Google Scholar - Bettelheim KA, Breadon A, Faiers MC, O'Farell SM, Shooter RA 1974 The origin of O-serotypes of Escherichia coli in babies after normal delivery. J Hyg (Lond) 72: 67–70
Article CAS Google Scholar - Fryklund B, Tullus K, Berglund B, Burman LG 1992 Importance of the environmental and the faecal flora of infants, nursing staff and parents as sources of Gramnegative bacteria colonizing newborns in three neonatal wards. Infection 20: 253–257
Article CAS PubMed Google Scholar - Lindberg E, Nowrouzian F, Adlerberth I, Wold A E 2000 Long-time persistence of superantigen-producing Staphylococcus aureus strains in the intestinal microflora of healthy infants. Pediatr Res 48: 741–747
Article CAS PubMed Google Scholar - Sears HJ, Brownlee I 1951 Further observations on the persistence of individual strains of Escherichia coli in the intestinal tract of man. J Bacteriol 63: 47–57
Google Scholar - Nowrouzian F, Wold AE, Adlerberth I 2001 P fimbriae and aerobactin as intestinal colonization factors for Escherichia coli in Pakistani infants. Epidemiol Infect 126: 19–23
CAS PubMed PubMed Central Google Scholar - Nowrouzian F, Adlerberth I, Wold AE 2001 P fimbriae, capsule and aerobactin characterize colonic resident Escherichia coli. Epidemiol Infect 126: 11–18
CAS PubMed PubMed Central Google Scholar - Wold AE, Caugant DA, Lidin-Janson G, Man PD, Svanborg C 1992 Resident colonic Escherichia coli strains frequently display uropathogenic characteristics. J Infect Dis 165: 46–52
Article CAS PubMed Google Scholar - Adlerberth I, Hanson LA, Svanborg C, Svennerholm AM, Nordgren S, Wold AE 1995 Adhesins of Escherichia coli associated with extraintestinal pathogenicity confer binding to colonic epithelial cells. Microb Pathog 18: 373–385
Article CAS PubMed Google Scholar - Strömberg N, Marklund BI, Lund B, Ilver D, Hamers A, Gaastra W, Karlsson KA, Normark S 1990 Host-specificity of uropathogenic Escherichia coli depends on differences in binding specificity to Galα1-4Gal-containing isoreceptors. EMBO J 9: 2001–2010
Article PubMed PubMed Central Google Scholar - Johnson JR 1998 PapG alleles among Escherichia coli strains causing urosepsis: associations with other bacterial characteristics and host compromise. Infect Immun 66: 4568–4571
CAS PubMed PubMed Central Google Scholar - Johanson M, Plos K, Marklund BI, Svanborg C 1993 Pap, papG and prsG DNA sequences in Escherichia coli from the fecal flora and the urinary tract. Microb Pathog 15: 121–129
Article CAS PubMed Google Scholar - Johnson JR, O'Bryan TT, Low DA, Ling G, Delavari P, Fasching C, Russo TA, Carlino U, Stell A L 2000 Evidence of commonality between canine and human extraintestinal pathogenic Escherichia coli strains that express papG allele III. Infect Immun 68: 3327–3336
Article CAS PubMed PubMed Central Google Scholar - Feria C, Machado J, Durate Correia J, Goncalves J, Gaastra W 2001 Distribution of papG alleles among uropathogenic Escherichia coli isolated from different species. FEMS Microbiol Lett 21: 205–208
Article Google Scholar - Gästrin B, Kallings LO, Marcetic A 1968 The survival time for different bacteria in various transport media. Acta Path Microbiol Scand 74: 371–380
Article PubMed Google Scholar - Kauffman F 1969 The Bacteriology of Enterobacteriaceae. Munksgaard, Copenhagen
- Ahrné S, Molin G, Ståhl S 1989 Plasmids in Lactobacillus strains isolated from meat and meat products. Syst Appl Microbiol 11: 320–325
Article Google Scholar - Nowrouzian F, Wold AE, Adlerberth I 2001 Computer-based analysis of RAPD (random amplified polymorphic DNA) fingerprints for typing of intestinal Escherichia coli. Mol Biol Today 2: 5–10
CAS Google Scholar - Le Bouguenec C, Archambaud M, Labigne A 1992 Rapid and specific detection of the pap, afa, and sfa adhesin-encoding operons in uropathogenic Escherichia coli strains by polymerase chain reaction. J Clin Microbiol 30: 1189–1193
CAS PubMed PubMed Central Google Scholar - Yamamoto S, Terai A, Yuri K, Kurazono H, Takeda Y, Yoshida O 1995 Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunol Med Microbiol 12: 85–90
Article CAS PubMed Google Scholar - Johnson JR, Brown JJ 1996 A novel multiply primed polymerase chain reaction assay for identification of variant papG genes encoding the Gal(α1-4)Gal-binding papG adhesins of Escherichia coli. J Infect Dis 173: 920–926
Article CAS PubMed Google Scholar - Johnson JR, Stapleton AE, Russo TA, Scheutz F, Brown JJ, Maslow JN 1997 Characteristics and prevalence within serogroup O4 of a J96-like clonal group of uropathogenic Escherichia coli O4:H5 containing the class I and class II alleles of papG. Infect Immun 65: 2153–2159
CAS PubMed PubMed Central Google Scholar - Sussman M 1997 Escherichia coli and human disease. In: Sussman M (ed) Escherichia coli Mechanism of Virulence. Cambridge University Press, Cambridge, U.K, pp 331–340
Google Scholar - Gareau FE, Mackel DC, Boring III, Payne FJ, Hammett FL 1959 The acquisition of fecal flora by infants from their mothers during birth. J Pediatr 54: 313–318
Article CAS PubMed Google Scholar - Gothefors L, Carlsson B, Ahlstedt S, Hanson LA, Winberg J 1976 Influence of maternal gut flora and colostral and cord serum antibodies on presence of Escherichia coli in faeces of the newborn infant. Acta Paediatr Scand 65: 225–232
Article CAS PubMed Google Scholar - Bettelheim KA, Teoh-Chan C, Chandler ME, O'Farrell SM, Rahamim L, Shaw EJ, Shooter RA 1974 Spread of Escherichia coli colonizing newborn babies and their mothers. J Hyg (Lond) 73: 383–387
Article CAS Google Scholar - Mata LJ, Urrutia JJ 1971 Intestinal colonization of breast-fed children in a rural area of low socioeconomic level. Ann NY Acad Sci 176: 93–109
Article Google Scholar - Adlerberth I, Carlsson B, de Man P, Jalil F, Khan SR, Larsson P, Mellander L, Svanborg C, Wold AE, Hanson LA 1991 Intestinal colonization with Enterobacteriaceae in Pakistani and Swedish hospital delivered infants. Acta Paediatr Scand 80: 602–610
Article CAS PubMed Google Scholar - Rotimi VO, Olowe SA, Ahmed I 1985 The development of bacterial flora of premature neonates. J Hyg (Lond) 94: 309–318
Article CAS Google Scholar - Bettelheim KA, Peddie BA, Chereshsky A 1983 The ecology of Escherichia coli in a maternity ward in Chirstchurch, New Zealand. Zentralbl Bakteriol Mikrobiol Hyg Ser B Umwelthyg Krankenhaushyg Arbeitshyg Praev Med 178: 389–393
CAS Google Scholar - Lundequist B, Nord CE, Winberg J 1985 The composition of the faecal microflora in breastfed and bottle fed infants from birth to eight weeks. Acta Paediatr Scand 74: 45–51
Article CAS PubMed Google Scholar - Adlerberth I, Jalil F, Carlsson B, Mellander L, Hanson LÅ, Larsson P, Khalil K, Wold AE 1998 High turnover rate of Escherichia coli strains in the intestinal flora of infants in Pakistan. Epidemiol Infect 121: 587–598
Article CAS PubMed PubMed Central Google Scholar - Kühn I, Tullus K, Möllby R 1986 Colonization and persistence of Escherichia coli phenotypes in the intestines of children aged 0 to 18 months. Infection 14: 7–12
Article PubMed Google Scholar - Mevissen-Verhage EA, Marcelis JH, Guinée PA, Verhoef J 1985 Effect of iron on serotypes and haemagglutination patterns of Escherichia coli in bottle-fed infants. Eur J Clin Microbiol 4: 570–574
Article CAS PubMed Google Scholar - van der Waaij D 1999 Microbial ecology of the intestinal microflora: influence of interactions with the host organism. In: Hanson LÅ, Yolken RH (eds) Probiotics, Other Nutritional Factors, and Intestinal Microflora. Lippincott-Raven, Philadelphia, pp 1–16
Google Scholar - Shroff KE, Meslin K, Cebra JJ 1995 Commensal enteric bacteria engender a selflimiting humoral mucosal immune response while permanently colonizing the gut. Infect Immun 63: 3904–3913
CAS PubMed PubMed Central Google Scholar - Mellander L, Carlsson B, Jalil F, Söderström T, Hanson LÅ 1985 Secretory IgA antibody response against Escherichia coli antigens in infants in relation to exposure. J Pediatr 107: 430–433
Article CAS PubMed Google Scholar - Freter R 1992 Factors affecting the microecology of the gut. In: Fuller R (ed) Probiotics—The Scientific Basis. Chapman 38 Hall, London, pp 111–144
Google Scholar - Adlerberth I, Hanson LÅ, Wold AE 1999 The ontogeny of the intestinal flora. In: Sanderson IR, Walker WA (eds) Development of the Gastrointestinal Tract. BC Decker Inc, Hamilton, Ontario, pp 279–292
Google Scholar - Stark PL, Lee A 1982 The microbial ecology of the large bowel of breast-fed and formula-fed infants during the first year of life. J Med Microbiol 15: 189–203
Article CAS PubMed Google Scholar - Duriez P, Clermont O, Bonacorsi S, Bingen E, Chaventré A, Elion J, Picard B, Denamur E 2001 Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology 147: 1671–1676
Article CAS PubMed Google Scholar - Ørskov F, Sorensen KB 1975 Escherichia coli serogroups in breast-fed and bottle-fed infants. Acta Pathol Microbiol Scand Sect B Microbiol 83: 25–30
Google Scholar - Tullus K, Kallenius G, Möllby R Fecal colonization with P-fimbriated E. coli between 0 and 18 months of age. Epidemiol Infect 100: 185–191
- Hesselmar B, Åberg N, Åberg B, Eriksson B, Björkstén B 1999 Does early exposure to cat or dog protect against later allergy development?. Clin Exp Allergy 29: 611–617
Article CAS PubMed Google Scholar - Bråbäck L, Kjellman N-I M, Sandin A, Björkstén B 2000 Atopy among schoolchildren in northern and southern Sweden in relation to pet ownership and early life events. Pediatr Allergy Immunol 12: 4–10
Article Google Scholar - Wold AE, Thorssén M, Hull S, Svanborg Edén C 1988 Attachment of Escherichia coli via mannose- or Galα1-4Galβ-containing receptors to human colonic epithelial cells. Infect Immun 56: 2531–2537
CAS PubMed PubMed Central Google Scholar - Wold AE, Mestecky J, Tomana M, Kobata A, Ohbayashi H, Endo T, Svanborg Edén C 1990 Secretory immunoglobulin A carries oligosaccharide receptors for Escherichia coli type 1 fimbrial lectin. Infect Immun 58: 3073–3077
CAS PubMed PubMed Central Google Scholar - Friman V, Adlerberth I, Conell H, Svanborg C, Hanson LÅ, Wold AE 1996 Decreased expression of mannose-specific adhesins by Escherichia coli in the colonic microflora of IgA-deficient individuals. Infect Immun 64: 2794–2798
CAS PubMed PubMed Central Google Scholar - Johnson JR 1991 Virulence factors in Escherichia coli urinary tract infection. Clin Microbiol Rev 4: 80–128
Article CAS PubMed PubMed Central Google Scholar - Smith HW, Linggood MA 1971 Observation on the pathogenic properties of the K88, Hly and Ent plasmids of Escherichia coli with particular reference to porcine diarrhoea. J Med Microbiol 4: 467–485
Article CAS PubMed Google Scholar - Krivan HC, Franklin DP, Wang W, Laux DC, Cohen PC 1992 Phosphatidylserine found in intestinal mucus serves as a sole source of carbon and nitrogen for Salmonellae and Escherichia coli. Infect Immun 60: 3943–3946
CAS PubMed PubMed Central Google Scholar - Hacker J, Blum-Oehler G, Mühldorfer I, Tschäpe H 1997 Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol 23: 1089–1097
Article CAS PubMed Google Scholar - Dobrindt U, Blum-Oehler G, Hartsch T, Gottschallk G, Ron EZ, Fünfstück R, Hacker J 2001 S-fimbria-encoding determinant _sfa_1 is located on pathogenicity island III536 of uropathogenic Escherichia coli strain 536. Infect Immun 69: 4248–4256
Article CAS PubMed PubMed Central Google Scholar - Bingen-Bidois M, Clermont O, Bonacorsi S, Terki M, Brahimi N, Loukil C, Barraud D, Bingen E 2002 Phylogenetic analysis and prevalence of urosepsis strains of Escherichia coli bearing pathogenicity island-like domains. Infect Immun 70: 3216–3226
Article CAS PubMed PubMed Central Google Scholar - Picard B, Sevali Garcia J, Gouriou S, Duriez P, Brahimi N, Bingen E, Elion J, Denamur E 1999 The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun 67: 546–553
CAS PubMed PubMed Central Google Scholar - Karch H, Schubert S, Zhang D, Zhang W, Schmidt H, Ölschläger T, Hacker J 1999 A genomic island, termed high-pathogenicity island, is present in certain non-O157 Shiga toxin-producing Escherichia coli clonal lineages. Infect Immun 67: 5994–6001
CAS PubMed PubMed Central Google Scholar
Author information
Authors and Affiliations
- Departments of Clinical Bacteriology, Sahlgrenska University Hospital, Göteborg, Sweden
Forough Nowrouzian, Agnes E Wold & Ingegerd Adlerberth - Departments of Pediatrics, Sahlgrenska University Hospital, Göteborg, Sweden
Bill Hesselmar, Robert Saalman, Inga-Lisa Strannegård & Nils Åberg
Authors
- Forough Nowrouzian
You can also search for this author inPubMed Google Scholar - Bill Hesselmar
You can also search for this author inPubMed Google Scholar - Robert Saalman
You can also search for this author inPubMed Google Scholar - Inga-Lisa Strannegård
You can also search for this author inPubMed Google Scholar - Nils Åberg
You can also search for this author inPubMed Google Scholar - Agnes E Wold
You can also search for this author inPubMed Google Scholar - Ingegerd Adlerberth
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toForough Nowrouzian.
Additional information
Supported by the Swedish Medical Research Council (K2001-06X-14072-01); the Swedish Foundation for Health Care Sciences and Allergy Research, the European Commission (QLK4-2000-00538); the Asthma and Allergy Foundation; the Ellen, Walter and Lennart Hesselman Foundation; and The Swedish Free Masonry Foundation.
Rights and permissions
About this article
Cite this article
Nowrouzian, F., Hesselmar, B., Saalman, R. et al. Escherichia coli in Infants' Intestinal Microflora: Colonization Rate, Strain Turnover, and Virulence Gene Carriage.Pediatr Res 54, 8–14 (2003). https://doi.org/10.1203/01.PDR.0000069843.20655.EE
- Received: 08 April 2002
- Accepted: 23 September 2002
- Issue Date: 01 July 2003
- DOI: https://doi.org/10.1203/01.PDR.0000069843.20655.EE