Estimated epidemiologic parameters and morbidity associated with pandemic H1N1 influenza (original) (raw)
Research
, Amy L. Greer, Michael Whelan, Anne-Luise Winter, Brenda Lee, Ping Yan, Jianhong Wu, Seyed Moghadas, David Buckeridge, Babak Pourbohloul and David N. Fisman
CMAJ February 09, 2010 182 (2) 131-136; DOI: https://doi.org/10.1503/cmaj.091807

Abstract
Background: In the face of an influenza pandemic, accurate estimates of epidemiologic parameters are required to help guide decision-making. We sought to estimate epidemiologic parameters for pandemic H1N1 influenza using data from initial reports of laboratory-confirmed cases.
Methods: We obtained data on laboratory-confirmed cases of pandemic H1N1 influenza reported in the province of Ontario, Canada, with dates of symptom onset between Apr. 13 and June 20, 2009. Incubation periods and duration of symptoms were estimated and fit to parametric distributions. We used competing-risk models to estimate risk of hospital admission and case-fatality rates. We used a Markov Chain Monte Carlo model to simulate disease transmission.
Results: The median incubation period was 4 days and the duration of symptoms was 7 days. Recovery was faster among patients less than 18 years old than among older patients (hazard ratio 1.23, 95% confidence interval 1.06–1.44). The risk of hospital admission was 4.5% (95% CI 3.8%–5.2%) and the case-fatality rate was 0.3% (95% CI 0.1%–0.5%). The risk of hospital admission was highest among patients less than 1 year old and those 65 years or older. Adults more than 50 years old comprised 7% of cases but accounted for 7 of 10 initial deaths (odds ratio 28.6, 95% confidence interval 7.3–111.2). From the simulation models, we estimated the following values (and 95% credible intervals): a mean basic reproductive number (_R_0, the number of new cases created by a single primary case in a susceptible population) of 1.31 (1.25–1.38), a mean latent period of 2.62 (2.28–3.12) days and a mean duration of infectiousness of 3.38 (2.06–4.69) days. From these values we estimated a serial interval (the average time from onset of infectiousness in a case to the onset of infectiousness in a person infected by that case) of 4–5 days.
Interpretation: The low estimates for _R_0 indicate that effective mitigation strategies may reduce the final epidemic impact of pandemic H1N1 influenza.
The emergence and global spread of pandemic H1N1 influenza led the World Health Organization to declare a pandemic on June 11, 2009. As the pandemic spreads, countries will need to make decisions about strategies to mitigate and control disease in the face of uncertainty.
For novel infectious diseases, accurate estimates of epidemiologic parameters can help guide decision-making. A key parameter for any new disease is the basic reproductive number (_R_0), defined as the average number of new cases created by a single primary case in a susceptible population. _R_0 affects the growth rate of an epidemic and the final number of infected people. It also informs the optimal choice of control strategies. Other key parameters that affect use of resources, disease burden and societal costs during a pandemic are duration of illness, rate of hospital admission and case-fatality rate. Early in an epidemic, the case-fatality rate may be underestimated because of the temporal lag between onset of infection and death; the delay between initial identification of a new case and death may lead to an apparent increase in deaths several weeks into an epidemic that is an artifact of the natural history of the disease.
We used data from initial reports of laboratory-confirmed pandemic H1N1 influenza to estimate epidemiologic parameters for pandemic H1N1 influenza. The parameters included _R_0, incubation period and duration of illness. We also estimated risk of hospital admission and case-fatality rates, which can be used to estimate the burden of illness likely to be associated with this disease.
Methods
Data collection
We collected individual-level data on laboratory-confirmed cases of pandemic H1N1 influenza in the province of Ontario, Canada, with a reported date of symptom onset between Apr. 13 and June 20, 2009. This cutoff was chosen because individual-level reporting of cases of pandemic H1N1 influenza in Ontario was stopped after this date.
We obtained the data from the province’s Integrated Public Health Information System. This system incorporates data provided by public health units engaged in the investigation of initial laboratory-confirmed cases of pandemic H1N1 influenza. For these cases, information was available on age, date of symptom onset and, for patients admitted to hospital, the date of admission and discharge and the outcome at discharge. Records included source of exposure if known (e.g., travel to Mexico for early cases), earliest possible exposure and, in a subset of cases, most recent possible exposure before onset of symptoms.
Estimation of incubation period
We restricted this analysis to case records with both an earliest and most recent date of exposure to an infectious case or locale, as well as a recorded date of symptom onset. The most likely date of exposure was estimated as the midpoint between the earliest and most recent dates of exposure. We assessed the goodness-of-fit of log-normal and Weibull distributions to observed distributions of incubation periods, with goodness-of-fit evaluated using graphic log–log plots.
Estimation of duration of symptoms, risk of hospital admission and case-fatality rate
We estimated the duration of symptoms (the interval between onset of illness and resolution of symptoms) using methods similar to those used to evaluate the period of incubation. We restricted our analysis to include cases with a recorded date of symptom onset before June 21, 2009, and a documented date of symptom resolution. We evaluated the association between age and time to resolution of symptoms using the log-rank test and through construction of Cox proportional hazards models.
Because the absence of a documented date of symptom resolution could signify either ongoing illness or missing data on symptom resolution, we performed sensitivity analyses in which missing dates of symptom resolution were randomly replaced based on the log-normal probability distribution of available durations of symptoms; we considered dates after June 21, 2009, as “censored” for the purposes of these analyses.
To estimate the risk of hospital admission and case-fatality rates, we used competing-risk survival models. 1 Such models can be used to estimate the predicted cumulative incidence of a given outcome over time if other “competing” outcomes do not occur. In the model evaluating risk of hospital admission, the competing outcomes were admission to hospital, death without admission to hospital (as occurred in a handful of cases), recovery without admission to hospital and “censored” (for cases in which none of the outcomes had occurred by the final date of evaluation). We used a “ratio of hazards” approach to account for temporal effects, allowing for the projection of likely long-term hospital stays and case-fatality rate. For a more detailed description of this approach and its derivation, see Appendix 1 (available at www.cmaj.ca/cgi/content/full/cmaj.091807/DC1).
Simulation of disease transmission
We used a Markov Chain Monte Carlo model to simulate disease transmission under varying assumptions around duration of infectiousness, latent period (infected, but noninfectious) and likelihood of asymptomatic infection. We assumed that people would become infected at a rate dependent on the number of infectious people in the Ontario population and the duration of infectiousness. We used data on history of travel to Mexico to classify imported cases. These cases were assumed to contribute to the transmission of infection; we used dates of symptom onset for these cases to model the initial introduction of pandemic H1N1 influenza into the Ontario population. The structure of the model is outlined in Appendix 2 (available at www.cmaj.ca/cgi/content/full/cmaj.091807/DC1).
We performed 1000 simulations in which daily reported counts of imported and locally acquired cases were assumed to Poisson-random variates. We generated best-fit values for _R_0, latent period and infectious period for each simulation using an optimization routine that minimized the sum-of-squares difference between model projections and the observed epidemic curve. From the simulations, we estimated mean parameter values and 95% credible intervals.
Results
A total of 3152 laboratory-confirmed cases of pandemic H1N1 influenza had reported dates of symptom onset between Apr. 13 and June 20, 2009. The mean age of the patients was 21.9 years (standard deviation 15.7). The age distribution is presented in Figure 1. From the total number of cases, we selected those with sufficient information to estimate incubation period, duration of symptoms, risk of hospital admission and case-fatality rate (Figure 2).
Incubation period
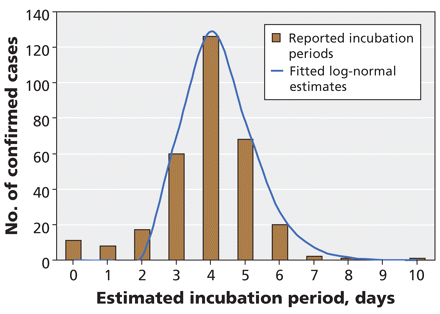
A total of 316 cases contained sufficient information on earliest exposure and disease onset to estimate incubation periods. The median time from exposure to symptom onset was 4 days (Figure 3). The distribution of estimated incubation periods followed a log-normal distribution (mean incubation 4.3 days, 95% CI 2.6–6.6 days).
Figure 3: Estimates of the incubation period (time from exposure to onset of symptoms) in 316 laboratory-confirmed cases of pandemic H1N1 influenza for whom these data were available. Estimates were calculated as the time between exposure date (midpoint of earliest and most recent exposure) and date of symptom onset. The blue curve represents the best-fit log-normal distribution of observed data.
Duration of symptoms
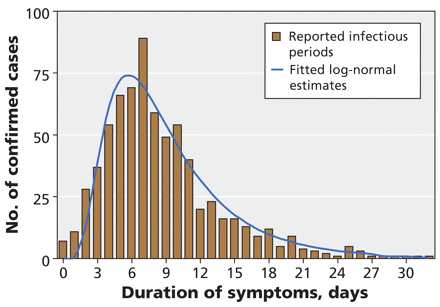
Of the 712 reported cases with documented resolution of symptoms, the median duration of symptoms was 7 days. The median duration was significantly shorter among patients aged less than 18 years (7 days) than among older patients (8 days) (hazard ratio 1.23, 95% confidence interval 1.06–1.44). Intervals between onset and resolution of symptoms followed a log-normal distribution (mean duration 9.3 days, 95% CI 2.6–24.2 days) (Figure 4).
Figure 4: Estimates of the duration of symptoms (time from onset to resolution of symptoms) in 712 laboratory-confirmed cases of pandemic H1N1 influenza for whom these data were available. The blue curve represents the best-fit log-normal distribution of observed data.
Risk of hospital admission and case-fatality rate
A total of 140 admissions to hospital and 10 deaths were recorded during the 10 weeks of observation. The estimated risk of hospital admission per case was 4.5% (95% CI 3.8%–5.2%). The estimated case-fatality rate was 0.3% (95% CI 0.1%–0.5%). In the competing-risk models used to adjust for recovery and death without admission to hospital, the cumulative risk of admission increased gradually during the first 5 days of illness, to about 3%. The risk was highest among infants (age < 1 year) and among elderly patients (age ≥ 65 years); adolescents were at decreased risk (Table 1). Although adults more than 50 years old comprised only 7% of cases, 7 of 10 deaths occurred in this age group (odds ratio 28.6, 95% CI 7.3–111.2). Using methods described above for the estimation of the case-fatality rate, we estimated an asymptomatic case-fatality rate of 0.002 (95% CI 0.0008–0.0076).
Table 1: Risk of hospital admission among 3152 laboratory-confirmed cases of pandemic H1N1 influenza, by age group
Model-based estimates of transmission parameters
We estimated a mean basic reproductive number (_R_0) of 1.31 (95% credible interval 1.25–1.38) (Figure 5); a mean latent period of 2.62 days (95% credible interval 2.28–3.12); and a mean duration of infectiousness of 3.38 days (95% credible interval 2.06–4.69). Using a commonly applied heuristic algorithm to estimate the serial interval 2 (the average time from onset of infectiousness in a case to the onset of infectiousness in a person infected by that case) based on the sum of the latent periods plus half the duration of infectiousness, we estimated the serial interval for pandemic H1N1 influenza to be 4–5 days. The range of _R_0 values would correspond with an overall attack rate (symptomatic and asymptomatic) of 20%–50% in the absence of intervention. 3
Figure 5: Estimates of the basic reproductive number (_R_0, number of new cases created by a single primary case in a susceptible population) in the province of Ontario during the wave of pandemic H1N1 influenza in the spring of 2009. Estimates were generated with the use of Markov Chain Monte Carlo simulation modelling. The mean estimate for _R_0 is 1.31 (95% credible interval 1.25–1.38).
Interpretation
We used initial reporting data on laboratory-confirmed cases of pandemic H1N1 influenza in the province of Ontario to estimate key epidemiologic parameters for pandemic H1N1 influenza. Because the 2009 influenza pandemic continues to evolve, these values are critical for planning and can be used to reduce some of the uncertainty around the health burden likely to be associated with this disease in the coming months. Our low estimate of 1.31 for the basic reproductive number (_R_0) is consistent with estimates from Mexico 4,5 (1.2 to 1.6) and the United States (1.7–1.8 after adjustment for increasing ascertainment of cases). 6 Our _R_0 estimate is also within a range where mitigation strategies, including use of antiviral drugs, social distancing and limited vaccination, might substantially reduce the final size of the epidemic. 7–11
Our estimated _R_0 may be lower than the true value for pandemic H1N1 influenza. Public concern related to the emerging pandemic may have changed population behaviour or medical practices, thereby reducing observed reproductive numbers. Furthermore, a striking feature of the current pandemic has been a concentration of cases in younger groups, which suggests the presence of pre-existing immunity in older adults. 12,13 The effective reproductive number in a partially immune population is lower than the basic reproductive number. 14 Despite reduced susceptibility, we have estimated that older people with pandemic H1N1 influenza are at increased risk of hospital admission and death, which emphasizes the need to characterize the relation between age and risk of infection early in a pandemic. 15 The discrepancy between age groups in susceptibility to infection and severity of illness may be an important factor limiting the burden of hospital admision and deaths in the current pandemic.
Our estimate of mean incubation period (time from infection to onset of symptoms) of 4.3 days is compatible with our model’s upper bound for latent period (time from infection to infectiousness) of 3.1 days, assuming a 1-day period of infectiousness before onset of symptoms. Both of these estimates are longer than is typical for seasonal influenza. 16 By contrast, our estimate of the duration of symptoms (median 7 days) is longer than our model-based estimate (mean 3.4 days). This difference could reflect effective isolation of cases or persistent symptoms that outlast infectiousness. However, our estimate of symptom duration is consistent with emerging data on shedding of the pandemic H1N1 influenza virus. 17 The long latent and infectious periods we have described resulted in longer estimated serial intervals for this disease than those previously generated by Fraser and colleagues using indirect methods. 4
The laboratory-confirmed cases we analyzed almost certainly represent a small subset of cases of pandemic H1N1 influenza during the period under study. We did not include infected people without symptoms or those with symptoms who did not undergo laboratory testing. Knowledge of the proportion of infected people who are actually identified as cases is needed to estimate the true risk of hospital admission or death among cases. 18 Estimates derived early in the current pandemic suggested that only 10% of people in developed countries received laboratory confirmation of pandemic H1N1 influenza. 18 More recent estimates are as low as 1.25%. 19 Thus, it may be reasonable to reduce our estimates of hospital admission and case-fatality rates by a factor of 10 or more in order to approximate risks among all people with pandemic H1N1 influenza. Serologic studies will be essential in developing more refined estimates of the true proportion of the population infected during the period under study.
Limitations
Our study has several limitations. First, our projection of hospital admissions and deaths among laboratory-confirmed cases, and with a limited number of outcomes, introduced wide confidence intervals in the risk estimates. As the pandemic progresses, we will be able to refine these values. However, our initial estimates provide some upper and lower bounds that may be useful for planning, if it is understood that laboratory-confirmed cases represent only a subset of all cases of pandemic H1N1 influenza in the community. Second, our estimates of the duration of symptoms may have been biased downward. Although we attempted to account for missing dates of symptom resolution by using statistical methods, some of the outcomes may have been censored because the patients had prolonged illness. Finally, not all hospital admissions or deaths attributable to influenza will be captured in surveillance data. For seasonal influenza, about 30% of related deaths have been predicted to occur outside of hospital, 20 and the burden of influenza difficult to assess. 21,22
Conclusion
On the basis of data from initial laboratory-confirmed cases of pandemic H1N1 influenza, our estimates show that the characteristics of this novel influenza virus do not differ substantially from those of seasonal influenza. However, when combined with high attack rates in younger groups, 13 there may be greater absolute numbers of hospital admissions and deaths than are observed in a typical influenza season. Enhanced surveillance during an influenza pandemic, compared with surveillance during seasonal influenza epidemics, could also contribute to such an effect. Our relatively low estimate of _R_0 (the number of new cases created by a single primary case in a susceptible population) indicates that effective use of mitigation strategies may substantially reduce the final size of the pandemic.
Footnotes
- Previously published at www.cmaj.ca
This article has been peer reviewed.
Competing interests: David Fisman has received research funds from Sanofi-Pasteur, which manufactures an influenza H1N1 vaccine used outside Canada. No competing interests declared by the other authors.
Contributors: Ashleigh Tuite, Amy Greer and David Fisman were involved in the study conception and design, the analysis and interpretation of data, and the drafting of the article. Michael Whelan was involved in the study conception and design, and the acquisition, analysis and interpretation of data. Anne-Luise Winter and Brenda Lee were involved in the acquisition, analysis and interpretation of data. Ping Yan, Jianhong Wu, Seyed Moghadas, David Buckeridge and Babak Pourbohloul were involved in the study design and the interpretation of data. All of the authors contributed to the critical revision of the manuscript for important intellectual content and approved the final version submitted for publication.
Funding: This work was supported by the Canadian Institutes of Health Research (CIHR) Pandemic Influenza Catalyst Grant Program, the Mathematics of Information Technology and Complex Systems (MITACS) Centre for Disease Modelling and the CIHR-supported Canadian Consortium for Pandemic Preparedness Modelling. Ashleigh Tuite receives support from the MITACS Accelerate Program through both MITACS funding and a generous matching contribution from the Ontario Agency for Health Protection and Promotion. Amy Greer was supported by a Post-doctoral Fellowship Award from the School of Graduate Studies at the University of Toronto during the initial phases of this work. David Fisman is supported by an Early Researcher Award from the Ontario Ministry of Research and Innovation and is supported by the Canadian Consortium for Pandemic Preparedness Modelling, MITACS and the CIHR. The opinions expressed in this article are those of the authors and do not represent official views or policy positions of the Ontario Agency for Health Protection and Promotion, the Ontario Ministry of Health and Long-Term Care or the National Research Council of Canada.
Acknowledgement: The authors acknowledge the assistance provided by Ms. Rachel Savage of the Ontario Agency for Health Protection and Promotion in the analysis and interpretation of data from the Integrated Public Health Information System.
REFERENCES
- 1.↵
Farley TM, Ali MM, Slaymaker E. Competing approaches to analysis of failure times with competing risks. Stat Med 2001;20:3601–10. - 2.↵
Lipsitch M, Cohen T, Cooper B, et al. Transmission dynamics and control of severe acute respiratory syndrome. Science 2003;300:1966–70. - 3.↵
Ma J, Earn DJ. Generality of the final size formula for an epidemic of a newly invading infectious disease. Bull Math Biol 2006;68:679–702. - 4.↵
Fraser C, Donnelly CA, Cauchemez S, et al. Pandemic potential of a strain of influenza A (H1N1): early findings. Science 2009;324:1557–61. - 5.↵
Pourbohloul B, Ahued A, Davoudi B, et al. Initial human transmission dynamics of the pandemic (H1N1) 2009 virus in North America. Influenza Other Respi Viruses 2009;3:215–22. - 6.↵
White LF, Wallinga J, Finelli L, et al. Estimation of the reproductive number and the serial interval in early phase of the 2009 influenza A/H1N1 pandemic in the USA. Influenza Other Respi Viruses 2009;3:267–76. - 7.↵
Longini IM Jr, Nizam A, Xu S, et al. Containing pandemic influenza at the source. Science 2005;309:1083–7. - Wu JT, Riley S, Fraser C, et al. Reducing the impact of the next influenza pandemic using household-based public health interventions. PLoS Med 2006; 3:e361.
- Vynnycky E, Pitman R, Siddiqui R, et al. Estimating the impact of childhood influenza vaccination programmes in England and Wales. Vaccine 2008;26: 5321–30.
- Germann TC, Kadau K, Longini IM Jr, et al. Mitigation strategies for pandemic influenza in the United States. Proc Natl Acad Sci U S A 2006;103:5935–40.
- 11.↵
Gojovic MZ, Sander B, Fisman D, et al. Modelling mitigation strategies for pandemic (H1N1) 2009. CMAJ 2009;181:673–80. - 12.↵
Hancock K, Veguilla V, Lu X, et al. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med 2009;361:1945–52. - 13.↵
Fisman DN, Savage R, Gubbay J, et al. Older age and a reduced likelihood of 2009 H1N1 virus infection. N Engl J Med 2009;361:2000–1. - 14.↵
- 15.↵
Miller MA, Viboud C, Olson DR, et al. Prioritization of influenza pandemic vaccination to minimize years of life lost. J Infect Dis 2008;198:305–11. - 16.↵
Heymann DL. Control of communicable diseases manual. Washington (DC): American Public Health Association; 2004. - 17.↵
De Serres G, Rouleau I, Hamelin M, et al. Shedding of novel 2009 pandemic H1N1 (nH1N1) virus at one week post illness onset. 49th Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC); 2009 Sept. 12–15; San Francisco (CA). Washington (DC); 2009. Abstract K-1918a. - 18.↵
Wilson N, Baker MG. The emerging influenza pandemic: estimating the case fatality ratio. Euro Surveill 2009;4. pii:19255. - 19.↵
Reed C, Angulo F, Swerdlow D, et al. Estimates of the prevalence of pandemic (H1N1) 2009, United States, April–July 2009. Emerg Infect Dis 2009. [Epub ahead of print]. Available: www.cdc.gov/eid/content/15/12/pdfs/09-1413.pdf (accessed 2009 Nov. 22). - 20.↵
Zhang X, Meltzer MI, Wortley PM. FluSurge —a tool to estimate demand for hospital services during the next pandemic influenza. Med Decis Making 2006;26: 617–23. - 21.↵
Glezen WP. Serious morbidity and mortality associated with influenza epidemics. Epidemiol Rev 1982;4:25–44. - 22.↵
Simonsen L, Clarke MJ, Williamson GD, et al. Schonberger: LB. The impact of influenza epidemics on mortality: introducing a severity index. Am J Public Health 1997;87:1944–50. - Paterson B, Durrheim DN, Tuyl F. Influenza: H1N1 goes to school. Science 2009; 325:1071–2.
- Chowell G, Miller MA, Viboud C. Seasonal influenza in the United States, France, and Australia: transmission and prospects for control. Epidemiol Infect 2008;136: 852–64.