Molecular architecture of the ribosome‐bound Hepatitis C Virus internal ribosomal entry site RNA (original) (raw)
Abstract
Internal ribosomal entry sites (IRESs) are structured _cis_‐acting RNAs that drive an alternative, cap‐independent translation initiation pathway. They are used by many viruses to hijack the translational machinery of the host cell. IRESs facilitate translation initiation by recruiting and actively manipulating the eukaryotic ribosome using only a subset of canonical initiation factor and IRES transacting factors. Here we present cryo‐EM reconstructions of the ribosome 80S‐ and 40S‐bound Hepatitis C Virus (HCV) IRES. The presence of four subpopulations for the 80S•HCV IRES complex reveals dynamic conformational modes of the complex. At a global resolution of 3.9 Å for the most stable complex, a derived atomic model reveals a complex fold of the IRES RNA and molecular details of its interaction with the ribosome. The comparison of obtained structures explains how a modular architecture facilitates mRNA loading and tRNA binding to the P‐site. This information provides the structural foundation for understanding the mechanism of HCV IRES RNA‐driven translation initiation.
Synopsis
A 3.9‐Å cryo‐EM structure of the human ribosome bound to the HCV IRES reveals how this viral RNA element hijacks the translational machinery of the host cell to facilitate replication.
- The cryo‐EM structure of the mammalian ribosomal 80S•HCV IRES complex has been solved at 3.9 Å.
- Large‐scale conformational changes such as inter‐subunit rotation and subunit rolling can be visualized.
- HCV IRES domain II is a highly dynamic element and promotes an open 40S subunit conformation by inducing a 40S head tilt via a wedge‐like mechanism.
- The structural basis for mRNA loading during HCV‐IRES‐driven internal initiation is provided.
Similar content being viewed by others
Introduction
Translation initiation is a key regulatory step in the control of gene expression. In eukaryotes recruitment, positioning, and activation of the protein synthesis machinery is highly dependent on the 5′ cap and 3′ poly(A) tail structures of the mRNA (Jackson et al, 2010). The complex pathway of canonical translation initiation includes an ATP‐dependent scanning mechanism and is orchestrated by the ordered action of about 12 initiation factors (eIFs) assembled from about 20 different polypeptides. However, some mRNAs are able to recruit the ribosome to the start codon in a more direct way. They can bypass cap binding and scanning and require only a reduced subset of eIFs, if any. This alternate pathway of internal initiation is driven instead by _cis_‐acting sequences in the 5′‐UTR of the mRNA called internal ribosomal entry sites (IRESs) (Hellen & de Breyne, 2007; Jackson et al, 2010). IRESs are used by many viral RNAs in order to hijack the translational apparatus of the host during infection. Furthermore, IRES elements in a subset of the cellular mRNAs may serve as regulatory elements to control, for example, mammalian gene expression during development (Xue et al, 2015).
The molecular mechanism of neither canonical nor internal translation initiation is fully understood. Moreover, there is no unique mechanism for internal initiation, and apparently, there are several molecular strategies to initiate translation as seen by the dependence of various IRES families on different sets of canonical eIFs and IRES transacting factors (Hellen & de Breyne, 2007; Jackson et al, 2010). However, the activity of several IRESs has been shown to depend on the ability of the IRES RNAs to adopt a tertiary fold, which in turn allows specific, complex interactions with the translational machinery. The finding that type 3 (HCV‐like) and type 4 (dicistrovirus intergenic region) IRESs can promote functionally important conformational changes in the ribosomal 40S subunit (Spahn et al, 2001; 2005) suggests that these IRES RNAs—similar to protein translation factors—are actively involved in the mechanisms of translation initiation and thus may be regarded as mRNA built‐in translation factors.
One of the prominent IRES family is constituted by the Hepatitis C Virus (HCV) IRES and related IRESs. The human‐pathogenic flavivirus HCV is a worldwide health threat and has chronically infected about 150 million individuals worldwide (Hajarizadeh et al, 2013). It is a main causative agent of liver diseases such as hepatocellular carcinoma and severe cirrhosis. HCV‐like IRESs are also found in closely related animal pathogenic pestiviruses (e.g. the classical swine fever virus, CSFV) as well as in some non‐related picornaviruses (e.g. simian sapelo virus 3, SSV‐3; and porcine teschovirus 1, PTV‐1) (Hellen & de Breyne, 2007). So far, this type of IRESs has been found in 24 virus genera (Asnani et al, 2015). Because the HCV IRES promotes translation initiation of its polyprotein, a key step during the HCV infection after the virus has entered its host cell, novel therapeutic approaches against HCV infections consider its IRES as a prime drug target (Jubin, 2001; Dibrov et al, 2014).
It has been shown by a variety of functional studies that the HCV and HCV‐like IRESs facilitate translation initiation throughout a complex pathway and control the formation of functional 48S‐like and 80S initiation complexes (Locker et al, 2007). The ability of the HCV IRES to bind with high affinity to the ribosomal 40S subunit placing the AUG initiation codon directly into the ribosomal P‐site is crucial for this pathway. Binding of the Met‐tRNAiMet•eIF2•GTP ternary complex and eIF3 to the binary 40S•HCV IRES complex leads to formation of the 48S‐like complex. Like in the late steps of canonical initiation, eIF5‐induced GTPase activity on eIF2 and dissociation of the factors is followed by eIF5B•GTP‐promoted subunit joining to yield elongation‐competent 80S complexes. In addition, there are even more basic pathways for the HCV IRES to accomplish translation initiation. Under stress conditions, the IRES facilitates an eIF2‐independent mode of initiation (Pestova et al, 2008; Terenin et al, 2008), and at elevated levels of divalent cations, the HCV IRES can operate even without the help of any initiation factor (Lancaster et al, 2006).
Knowledge of the molecular structure of HCV and HCV‐like IRES elements is of utmost importance not only for understanding the mechanism of internal initiation but also to support the development of novel antiviral inhibitors. The HCV IRES RNA contains 341 nucleotides, and the tertiary fold is formed by secondary structure domains II and III of the 5′ UTR. Domain I is not important for the IRES activity, and domain IV containing the AUG start codon unfolds upon binding to the 40S subunit (Reynolds et al, 1996; Pestova et al, 1998). Previous low‐ and subnanometer‐resolution cryo‐EM reconstructions of ribosomal 40S•HCV IRES (Spahn et al, 2001), 80S•HCV IRES (Boehringer et al, 2005), 80S•HCV IRES•Met‐tRNAi•eIF5B•GMPPNP (Yamamoto et al, 2014), and 40S•CSFV IRES (lacking domain II)•eIF3 (Hashem et al, 2013) complexes have shown that the HCV IRES forms an elongated structure. IRES domain III binds at the solvent side of the 40S subunit, whereas IRES domain II loops out toward the 60S side of the 40S head and reaches into the E‐site region. The structure of 80S•HCV IRES•Met‐tRNAi•eIF5B•GMPPNP showed the tip of domain II released from its canonical position interacting with the elbow of Met‐tRNAi during 60S subunit joining (Yamamoto et al, 2014). Moreover, several NMR and X‐ray studies provide high‐resolution information for many individual HCV IRES subdomains. In the absence of a high‐resolution structure of the full‐length HCV IRES, fitting these structure pieces into the intermediate‐resolution cryo‐EM maps allowed modeling of the HCV IRES in the ribosome‐bound form and a first analysis of the contacts to the ribosome (Boehringer et al, 2005; Berry et al, 2011). However, as this “divide and conquer” approach always leaves certain ambiguities, structural information at higher resolution is required to understand the molecular mechanism of HCV IRES‐driven translation initiation. Capitalizing on recent technological breakthrough in cryo‐electron microscopy (cryo‐EM), we present here an atomic model of the ribosome‐bound HCV IRES RNA based on a reconstruction of a binary 80S•HCV IRES complex to 3.9 Å resolution. Furthermore, our analysis reveals large‐scale conformational modes of 40S subunit within the 80S complex and suggests how domain II of the IRES facilitates mRNA loading and P‐site tRNA binding by using a wedge‐like mechanism to enforce 40S head tilting.
Results
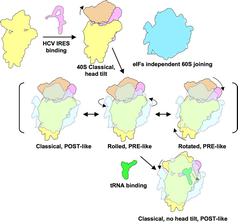
Ribosomal subunit rearrangements within the 80S•HCV IRES complex
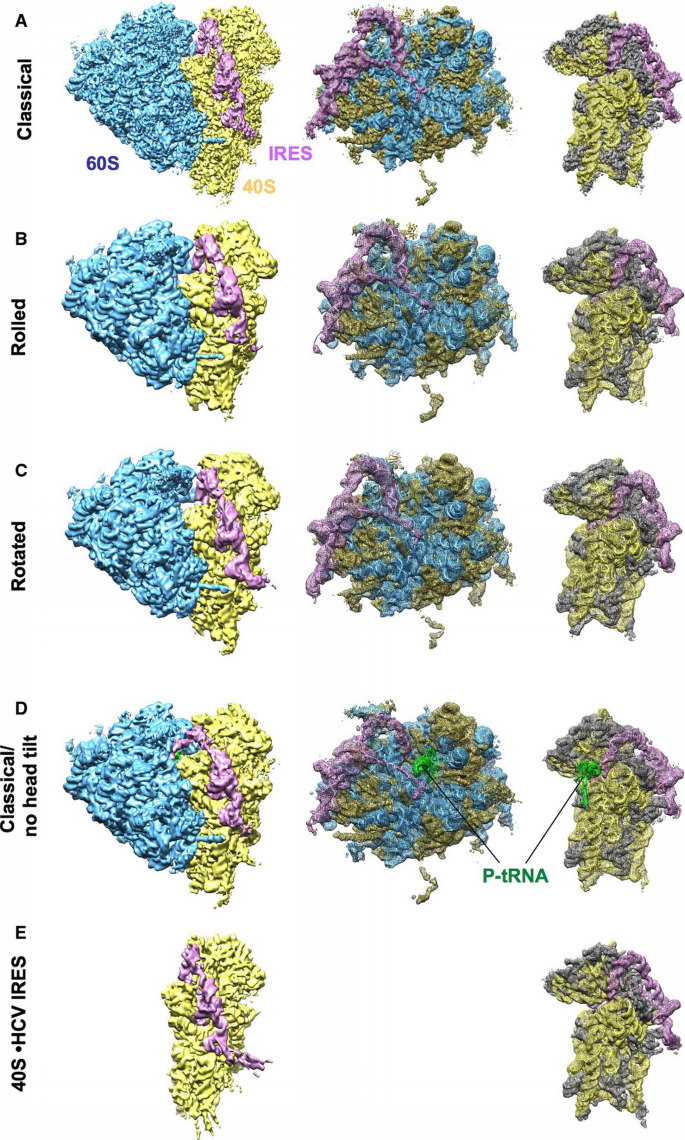
In order to reduce the potential compositional heterogeneity of complexes for cryo‐EM experiments, we made use of the finding that at elevated levels of divalent cations, no initiation factors are required for the formation of translation‐competent 80S•HCV IRES complexes (Lancaster et al, 2006). Accordingly, eIF‐free, binary 80S•HCV IRES complexes from purified rabbit ribosomal subunits and purified IRES RNA were assembled in the presence of Ca2+in vitro. Nevertheless, multiparticle classification techniques (Loerke et al, 2010) of the cryo‐EM data revealed considerable conformational heterogeneity (Fig EV1). Besides the predominant subpopulation that we could refine to high resolution, a first tier of classification resulted in two additional subpopulations of the 80S•HCV IRES complex with clear density for the HCV IRES (Fig 1A–C). In all three subpopulations and in general agreement with previous low‐resolution cryo‐EM studies (Spahn et al, 2001; Boehringer et al, 2005), the HCV IRES forms an elongated structure at the solvent side of the 40S subunit with IRES domain II folding back toward the E‐site region. However, the three subpopulations are distinguished by the ribosomal subunit arrangement as the 40S subunit together with the HCV IRES undergoes rotational movements relative to the 60S subunit (Fig 2A).
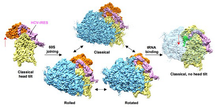
Figure 1
Cryo‐EM reconstructions of the 80S•HCV IRES and 40S•HCV IRES complexes
A–C Classical (A), rolled (B), and rotated (C) subpopulations of the 80S•HCV IRES complex.
D Subpopulation of the 80S•HCV IRES complex containing P‐site tRNA. The 80S is in the classical configuration and the head tilt is not present.
E 40S•HCV IRES. The cryo‐EM map was filtered according to resolution.
Data information: Left column shows an overview of the final cryo‐EM reconstructions with the HCV IRES (pink), 40S subunit (yellow), 60S subunit (blue), and tRNA (green). Middle and right columns show mesh representation of the maps and docked models of the 60S and 40S parts, respectively. 28S‐5S‐5.8S rRNA (blue), 60S proteins (gold), 18S rRNA (yellow), 40S proteins (dark gray), HCV IRES (pink), tRNA (green).
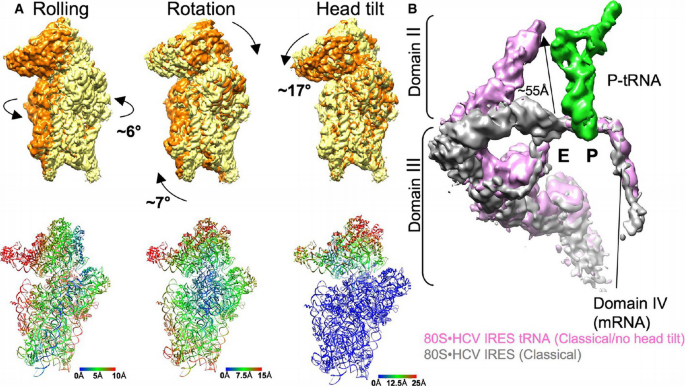
Figure 2
Analysis of intersubunit rearrangements and movement of the HCV IRES domain II
A. Comparison of 40S subunit positions in rolled, rotated, classical/no head tilt (orange), and classical 80S•HCV IRES complex (yellow) in common 60S alignment. The cryo‐EM map of the classical 80S•HCV IRES complex was filtered to 6 Å. Arrows indicate the direction of movement during transition between two different states. The distance changes in the 40S subunit positions resulting from the rigid body transformation are color‐coded in Å units.
B. Comparison of the HCV IRES in classical 80S•HCV IRES complex (gray) and upon P‐tRNA binding (classical, no head tilt; purple) in a common 40S body alignment. The distance of the movement of domain II is indicated with an arrow and Å units.
With respect to the relative arrangement of 40S and 60S subunits, the major subpopulation (classical) is related to the elongating 80S ribosome in the post‐translocational (POST) state (Budkevich et al, 2014). In addition, we found two conformational states (Rolled and Rotated) that bear resemblance to the classical and the rotated pre‐translocational (PRE) states, respectively (Budkevich et al, 2011). Accordingly, subunit rolling, which is characteristic for the classical PRE state in eukaryotes, and the orthogonal universal intersubunit rotation are present also in the factor‐free 80S•HCV IRES complex. In line with the metastable energy landscape view for translation (Munro et al, 2009; Behrmann et al, 2015), we attribute the presence of conformational modes to the intrinsically dynamic nature of the ribosome.
40S head tilt reversed by P‐site bound tRNA
In an attempt to further improve the resolution of the major subpopulation, we performed a second tier of classification. This resulted not only in an improved density for the major subpopulation but surprisingly also in a fourth subpopulation that contains additional density in the ribosomal intersubunit space (Fig 1D). Based on shape, size, and binding position, we interpret this density to represent P‐site bound tRNA (Fig EV2). Because we did not include tRNA when assembling the 80S•HCV IRES complex, this density has to stem from endogenous tRNA, co‐purified with the ribosomal subunits. Preparation of ribosomes often contain co‐purified tRNA that is usually located in the ribosomal E‐site (Cate et al, 1999; Anger et al, 2013; Khatter et al, 2015). The present P‐site location thus may reflect on the capability of the HCV IRES to direct initiator tRNA to the ribosomal P‐site during internal initiation.
The presence of P‐site tRNA density goes along with large‐scale conformational changes in the ribosomal 40S subunit and the HCV IRES. Previously, it was shown by cryo‐EM of 40S•HCV IRES complexes at ~20 Å resolution that domain II of the HCV IRES actively manipulates the conformation of the ribosomal 40S subunit (Spahn et al, 2001). In the present cryo‐EM reconstruction of a binary 40S•HCV IRES complex (Fig 1E), we can confirm this reorientation of the 40S head relative to the 40S body/platform at the improved resolution of 6.7 Å (Figs EV3 and EV4; Table EV1). Comparison with the three subpopulations of the binary 80S•HCV IRES complexes (Fig 1A–C) shows essentially the same 17° 40S head tilt upon association with the 60S subunit. Within the P‐site tRNA containing 80S•HCV IRES complex, however, the head tilt is reversed (Fig 2A) and the 40S subunit adopts not only the overall position but also the conformation of the 40S subunit within the elongating POST ribosome (Budkevich et al, 2014). Interestingly, the reversal of the 40S head tilt happens in conjunction with a pronounced movement of HCV IRES domain II by ~55 Å (Fig 2B). It detaches from its binding position on the 40S head and moves toward the 60S subunit to occupy a position in the E‐site region.
Structure of the ribosome‐bound HCV IRES RNA
The final reconstruction of the largest subpopulation was obtained from a subset of 171,820 particle images (39% of the full data set) and reached a resolution of 3.9 Å (Figs 1A, EV1 and EV3, Table EV1). In agreement with this resolution estimation, the core of the 80S ribosomes shows features expected at near‐atomic resolution (Fig EV4A). However, the density corresponding to the HCV IRES is of somewhat lower resolution. It is best defined in regions that are in close contact with the ribosome, for example, the central core of the IRES at the back of the platform or the apical part of domain II in the intersubunit space (Fig EV4A). It is therefore likely that dynamic properties of the IRES, that is, local flexibility, are the main resolution limiting factor. Nevertheless, the clear improvement in resolution compared to previous cryo‐EM reconstructions of the ribosome‐bound HCV IRES at intermediate (Spahn et al, 2001; Filbin et al, 2013) and also subnanometer resolution (Yamamoto et al, 2014) greatly aided modeling of the IRES RNA (Fig 3A and B).
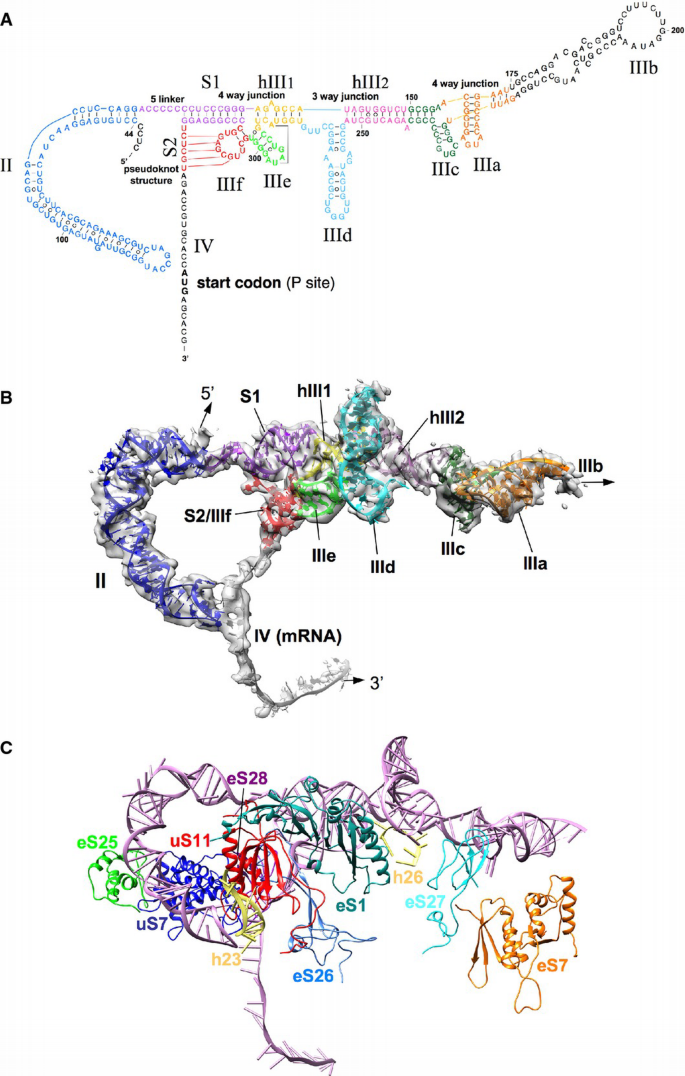
Figure 3
Structural model of the HCV IRES bound to the 80S ribosome
A. Adopted secondary structure of the IRES representing the tertiary fold.
B. Atomic model of the HCV IRES RNA. IRES domains are presented as colored ribbons, overlaid with experimental IRES density (gray, semitransparent).
C. Molecular model of the HCV IRES (pink) with ribosomal interaction sites (colored proteins and helices).
Structural modeling of the IRES was aided by X‐ray/NMR structures for several HCV IRES subdomains (Lukavsky, 2009; Berry et al, 2011), and the present map allows a more accurate placement of these structures of individual subdomains compared to previous placements into low‐resolution cryo‐EM maps (Boehringer et al, 2005; Berry et al, 2011). Moreover, fitting of some of these HCV IRES subdomain structures required partial remodeling indicating that conformational changes can occur in the context of the complete IRES RNA or due to interactions with the ribosome. For example, the NMR structures of HCV IRES subdomains IIa and IIb (Lukavsky et al, 2003) fitted readily into the respective cryo‐EM density (Fig EV5). However, compared to the NMR structure of the complete domain II in solution (Lukavsky et al, 2003), the curvature for the ribosome‐bound IRES domain II has increased indicating a conformational change at the junction between the two subdomains upon ribosome binding (Fig EV5).
According to the secondary structure of the HCV IRES (Kieft et al, 2002; Hellen & de Breyne, 2007), the basal end of domain II is followed by a short 5‐nucleotide linker and the 9‐base pair stem I (S1) (Fig 3A and B). S1 together with subdomains IIIe, IIIf, and helix III1 adopts an unusual four‐way junction, because the loop region of IIIf is involved in basepairing interactions (stem II; S2) with a RNA strand of the 3′ region of the IRES to build a pseudoknot structure (Berry et al, 2011). The crystal structure of this complex RNA module (Berry et al, 2011), which includes a predicted tertiary interaction between U297 of IIIe and A288 of hIII1 (Fig 3A) (Easton et al, 2009), could be readily incorporated into our overall model (Figs 3B and EV5). Emanating from (and directed through) S2 of the HCV IRES central domain, domain IV passes through the mRNA binding channel placing the AUG in the ribosomal P‐site (Figs 2B and 3B). As suggested previously, unwinding of domain IV upon ribosomal binding is necessary for functional translation initiation via the HCV IRES (Honda et al, 1996; Filbin & Kieft, 2011) and we can trace ~27 nucleotides of single‐stranded RNA in the mRNA groove corresponding to nucleotides 330–356 of the viral RNA (Fig 3B). In line with a previous prediction (Spahn et al, 2001), we found a tertiary interaction of domain IV with the apical loop of domain II in the ribosomal E‐site region (Fig 2B).
On the other side of the central four‐way junction, helix hIII1 leads into a three‐way junction between hIII1, hIII2, and subdomain IIId. Helices hIII1 and hIII2 are coaxially stacked on each other and subdomain IIId adopts an approximately perpendicular orientation (Fig 3B). Like domain II, the NMR structure of isolated subdomain IIId (PDB 1FQZ) (Klinck et al, 2000) provided a good starting point for our model of the complete ribosome‐bound HCV IRES. The subdomain IIId contains an internal loop motif characteristic of the rRNA α‐sarcin/ricin loop. In contrast, the related CSFV IRES possesses a IIId stem composed of standard Watson–Crick base pairs only (Hashem et al, 2013). The apical loop UUGGGU of isolated IIId folds into a trinucleotide loop closed by a transwobble U‐G base pair (Klinck et al, 2000; Lukavsky et al, 2000). However, the loop conformation had to be changed to account for a conformational change that occurs upon interaction with the 18S rRNA (see below). The single‐stranded CUUG connecting the basal side of subdomain IIId to the three‐way junction was modeled as tetra loop with the closing C:G pair stacked onto IIId (Fig EV5).
Helix hIII2 together with subdomains IIIc, IIIa, and IIIb forms the IIIabc four‐way junction (Fig 3B). The crystal structure of the isolated IIIabc junction was solved and found to resemble a cH‐type four‐way junction with two coaxially stacked helices and the continuous strands running in parallel direction (PDB 1KH6)(Kieft et al, 2002). Interestingly, our density is not compatible with the parallel cH type. In the ribosome‐bound form the IIIabc junction appears to adopt an L‐shaped cL‐type fold (Laing & Schlick, 2009) with the two helices IIIa/IIIb coaxially stacked and IIIc/hIII2 being arranged perpendicular to each other and with interactions between the loops of IIIa and IIIc (Figs 3B and EV5). The X‐ray structure of the isolated IIIabc junction is based on a crystal induced dimer, in which the loop of IIIc was eliminated (Kieft et al, 2002). As the interactions of loop IIIc with loop IIIa and also the 40S subunit appear critical for the observed cL‐type fold of the ribosome‐bound IIIabc junction, this may explain the discrepancy between both structures. We do not observe major differences between the present 40S•HCV IRES complex (Fig 1E) and the binary 80S•HCV IRES complex (Fig 1A) with respect to overall IRES structure. Thus, our analysis is not compatible with a proposal made based on a low‐resolution cryo‐EM map of an HCV IRES‐80S complex that the IIIabc junction adopts an antiparallel H‐type conformation and that a functionally important transition from the cH‐ to the H‐type occurs upon subunit association (Boehringer et al, 2005).
After the IIIabc junction, our cryo‐EM map shows density for only a small part of subdomain IIIb, probably due to flexibility (Fig 3B). Therefore, we refrained from modeling IIIb and our model is missing about 50 nucleotides in this area. Nevertheless, the visible part of this mostly helical subdomain suggests that IIIb sticks out far from the 40S subunit and into solution, in agreement with the cryo‐EM reconstructions of the 40S•CSFV IRES complex (Hashem et al, 2013).
Interaction partners of the HCV IRES RNA with the ribosome
Facilitated by the recent atomic models for the human 80S ribosome based on near‐atomic resolution cryo‐EM reconstructions of the porcine (Voorhees et al, 2014) or the human 80S ribosome (Behrmann et al, 2015; Khatter et al, 2015), our present cryo‐EM map allows an interpretation of the molecular interactions between the HCV IRES RNA and the ribosome. According to biochemical assays, mainly ribosomal proteins have been implicated as interaction partner of the HCV IRES RNA (Fukushi et al, 2001; Otto et al, 2002; Laletina et al, 2006; Babaylova et al, 2009). Also the re‐analysis of low‐resolution maps of 80S•HCV IRES and 40S•HCV IRES complexes suggested interactions with uS7, eS25, eS26, eS1, and eS27 (Joseph et al, 2014). In general agreement, we observe interactions with several ribosomal proteins throughout the various contact regions (Fig 3C, Table EV2). eS7 and eS27 facilitate binding of the IIIabc junction at IIIa and IIIc, respectively, in agreement with previous biochemical experiments (Malygin et al, 2013b) (Fig EV6).
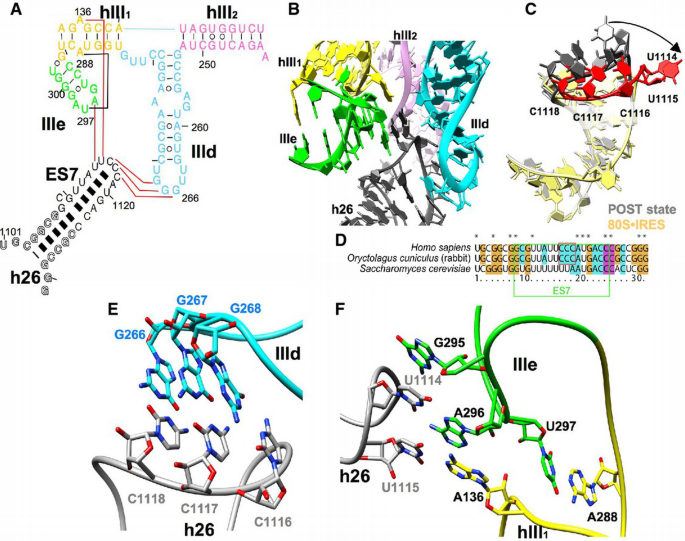
However, the central interaction network is based on complex RNA–RNA interactions between subdomains IIId and IIIe of the central core of the HCV IRES and the apical loop of expansion segment 7 (ES7 of helix h26) of 18S rRNA (Fig 4A–E). Subdomains IIId and IIIe have been implicated as major determinants for IRES binding by a large body of evidence (Kolupaeva et al, 2000; Kieft et al, 2001). Rationalizing recent chemical probing and mutagenesis studies (Kolupaeva et al, 2000; Malygin et al, 2013a; Matsuda & Mauro, 2014), our model shows a three‐nucleotide mini‐helix between the 266GGG268 sequence in the apical loop of subdomain IIId with the complementary 1116CCC1118 sequence in the loop of ES7 (Fig 4A and E). Formation of this mini‐helix requires conformational changes in both the apical loop of IIId as defined by the NMR structure (Klinck et al, 2000; Lukavsky et al, 2000) and ES7.
Figure 4
The interaction core between the HCV IRES and ES7 in helix 26 of the 18S rRNA
A. The secondary structure of the HCV IRES domain III (hIII1, hIII2, IIIe, and IIId) and helix 26 (h26) from the 18S rRNA. Colors of HCV IRES domains are the same as in Fig 3. Interaction between the HCV IRES and 18S rRNA are indicated as red lines, and a tertiary IRES interaction is indicated as a black line.
B. The interaction core between IIId, IIIe, hIII1 of HCV IRES (colored as in A) and expansion segment 7 (ES7) of h26 (dark gray).
C. The rearrangement of h26 (red) upon binding of HCV IRES. The 80S•HCV IRES complex (yellow) is compared to the elongating POST state (gray; PDB 5Aj0) (Behrmann et al, 2015). The loop closing the helix is highlighted in red and dark gray.
D. Primary sequence comparison of helix 26. ES7 is boxed in green. The three apical cytosine bases in the loop region of human and rabbit h26 are boxed in red.
E. Close‐up on the interactions between GGG266–268 of IIId (cyan) and CCC1116–1118 of ES7 (gray).
F. Close‐up on the interactions between G295, A296, and U297 of IIIe (green); A136 and A288 of hIII1 (yellow); and U1114 and U1115 of ES7 (gray).
The conformational change in the ES7 loop also repositions the adjacent 1114UU1115 (Fig 4C and F) to facilitate further contacts that appear to involve A296 within the loop of IRES subdomain IIIe and the bulged out A136 of helix hIII1. These interactions may be further stabilized by a stacking interaction of G295 (IIIe) with U1114 of ES7. The complimentary ES7 sequence 1116CCC1118 to HCV IRES domain IIId is well conserved only in higher eukaryotes (Fig 4D). Both nucleotides of the A136:A288 purine–purine mismatch within hIII1 are involved in tertiary/quaternary interactions. U297 of loop IIIe forms a preformed tertiary base pair with A288 of hIII1 (Easton et al, 2009; Berry et al, 2011), which leads to a position of the flipped out base of A136 next to A296. Simultaneous interactions of ES7 U1115 and U1114 with A136 and A296, respectively, provide an additional bridge between hIII1 and IIIe (Fig 4F).
In agreement with low‐resolution cryo‐EM maps (Spahn et al, 2001; Boehringer et al, 2005), domain II of the HCV IRES interacts with the 40S head. Interestingly, these interactions depend on the tilt movement of the 40S head relative to the body/platform domains (Fig 5A left panel). A common 40S body alignment of the major 80S•HCV IRES subpopulation and the canonical POST state (Behrmann et al, 2015) reveals that uS7 as positioned by the untilted 40S head of the POST state would clash with the apical loop of domain II as observed in the 80S•HCV IRES complex (Fig 5A right panel). This suggests that displacement of uS7 by the apical loop of domain II facilitates the head tilt movement and rationalizes previous observations that mutant HCV IRES RNAs with a deletion of the whole domain II or mutations at tip of domain II are defective in inducing the head tilt movement (Spahn et al, 2001; Filbin et al, 2013).
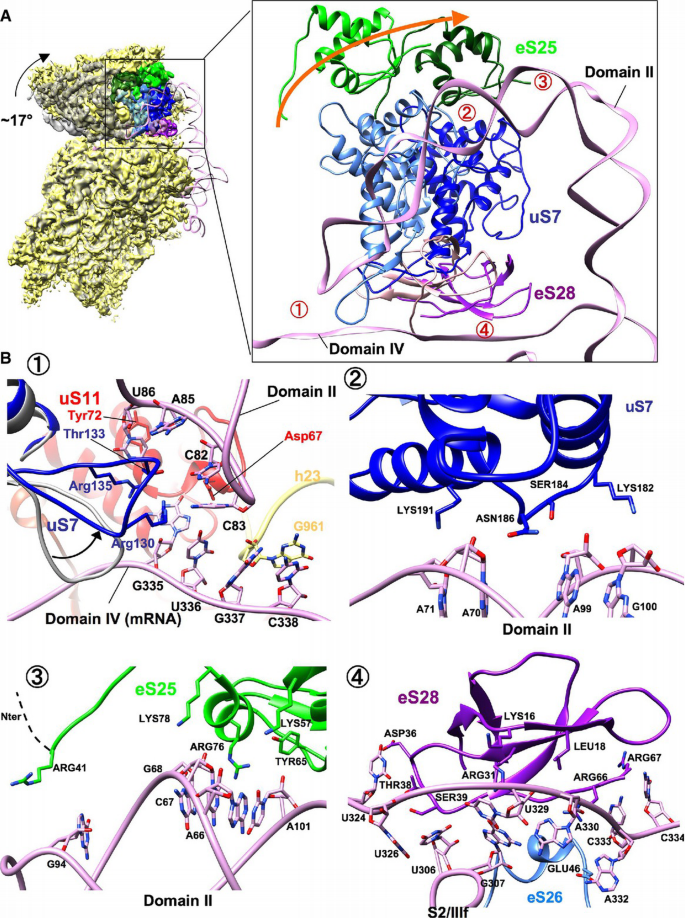
Figure 5
HCV IRES domain II displaces uS7 and induces the head tilt
A. Comparison of the 40S subunit from classical 80S•HCV IRES complex (yellow) and canonical POST state (gray; EMD 2875) (Behrmann et al, 2015) in common 40S body alignment. The arrow indicates the direction of the head tilt movement. Ribosomal proteins of the 40S head (uS7, eS25 and eS28) that interact with the HCV IRES domains II and IV (pink) on the 40S head are illuminated with blue, green, and purple, respectively. The positions of the proteins in the canonical POST state are illuminated in lighter color. The numbers indicate interaction sites of HCV IRES and the 40S subunit.
B. Close‐up view on the interaction site of HCV IRES and the 40S head region. Numbers correspond to red labels in (A). The rearrangement of the uS7 loop (blue) compared to the classical 80S•HCV IRES complex (gray) is indicated. The arrow indicates the direction of the rearrangement of the uS7 loop. B‐1 and B‐4: intersubunit view from 40S subunit. B‐2 and B‐3: intersubunit view from 60S subunit.
Rationalizing previous biochemical studies (Fukushi et al, 2001; Otto et al, 2002; Babaylova et al, 2009; Bhat et al, 2015), the functionally important IRES domain II interacts with the proteins uS7 and eS25 in the 40S head (Fig 5A right panel), as well as with uS11 in the platform region (Fig 5B‐1). In our model, the interactions of the middle region of domain II with uS7 and eS25 involve several positively charged Lys and Arg residues. Furthermore, there is a tentative contact between Arg41 of the long N‐terminal extension of eS25 with G94 of IRES domain II (Fig 5B‐2 and ‐3). The more N‐terminal part of eS25 appears disordered in our map. The apical loop region of domain II interacts with a β‐hairpin loop of uS7 that is reoriented compared to the POST state (Behrmann et al, 2015) and uS11 (Fig 5B‐1; Table EV2). Furthermore, C83 at the tip of domain II is seen to contact the single‐strand region of domain IV around G335 and U336 (Fig 5B‐1). According to a deletion analysis of the β‐hairpin loop of uS7 in bacteria, deletion of this loop increases frameshift due to most likely loosing E‐tRNA after translocation (Devaraj et al, 2009). For eukaryotes, loop mutations in uS7 lowered initiation fidelity (Visweswaraiah et al, 2015). Thus, the β‐hairpin in loop of uS7 may participate in keeping the reading frame during HCV IRES‐driven initiation. The deletion of domain II loses the activity completely; however, substitution or deletion of tip of domain II reduce the activity to around 30–50% (Odreman‐Macchioli et al, 2001; Kalliampakou et al, 2002; Filbin & Kieft, 2011). The interaction of the tip of domain II with the mRNA part of domain IV is not necessarily sequence specific.
In addition to h23, the single‐stranded domain IV is in close neighborhood to proteins uS11, eS26, and eS28 (Fig 5B‐1 and ‐4), which is in excellent agreement to the surrounding of the E‐site codon of canonical mRNA in 48S or 80S initiation complexes (Pisarev et al, 2008). Moreover, the β‐sheet structure of eS28 lines up to the IRES S2/IIIf region at nucleotides 324–334 (Fig 5B‐4) and next to eS26 and uS11, protein eS1 is seen to contact the hairpin structure IIIe of the central four‐way junction (Fig EV6). The observed interaction proteins in our work are in overall agreement with the re‐analysis of low‐resolution maps of 80S and 40S•HCV IRES complex suggesting interactions with uS7, eS25, eS26, eS1, and eS27 (Joseph et al, 2014).
Compared to the HCV IRES, the CSFV IRES has a shorter domain II and possesses the additional subdomain IIId2, which—according to data from the closely related Seneca Valley virus—does not contribute to activity (Willcocks et al, 2011). Nevertheless, both IRESs share a common domain architecture and bind to essentially the same binding site on the ribosomal 40S subunit (Hashem et al, 2013). However, because the reported CSFV IRES structure lacks domain II, the difference in domain II remains to be elucidated.
Discussion
Structural basis for hijacking of the ribosome by the HCV IRES
Our analysis suggests how a modular architecture of the HCV IRES facilitates internal initiation by interacting with ribosomal RNA and proteins (Figs 4 and 5, Table EV2). Domain III as defined by the secondary structure adopts a bipartic fold to recruit the 40S subunit (Fig 3). The core of the HCV IRES adopts a complex fold around the central four‐way junction and the three‐way junction between hIII1, hIII2, and subdomain IIId (Fig 4B). This tertiary RNA domain binds to the upper part of the 40S platform. The second tertiary domain formed by the IIIabc four‐way junction establishes separate contacts at the lower part of the platform. This is in excellent agreement with previous biochemical data showing that secondary structure domain III provides essentially all the binding affinity of the HCV IRES for the ribosomal 40S subunit and that especially IIIe, IIId, IIIa, and IIIc are major determinants of IRES binding (Kieft et al, 1999, 2001) (Fig 4).
Interestingly, complete folding of the larger, central RNA tertiary domain appears to require binding to the ribosome. The loop of ES7 of h26 in 18S rRNA intercalates between IRES stem loops IIId and IIIe and helix hIII1 and facilitates a network of interactions between these three elements (Figs 3 and 4). The directly adjacent 1116CCC1118 of ES7 in turn extends the interaction network to the IIId loop. Small‐angle X‐ray scattering data have suggested that the HCV IRES in solution is quite mobile and consists of rigid parts that move relative to each other (Perard et al, 2013). Accordingly, the presented network of RNA–RNA interactions may be crucial to compact and rigidify the IRES upon binding. Interestingly, these three cytosine bases can be used for other viral RNA re‐initiation as well (Luttermann & Meyers, 2009; Zinoviev et al, 2015). The usage of ES7 and adaptation of viral RNA is wider than anticipated.
Binding of the IIIabc four‐way junction to the lower part of the platform constrains the position of subdomain IIIb. However, our cryo‐EM map shows fragmented density for subdomain IIIb, indicating flexibility, and thus, our model lacks about 50 nucleotides (Fig 3B). Nevertheless, the trajectory inferred from the map suggests that IIIb sticks out far into solution. Previously, a IIIb position much closer to the 40S subunit and overlapping with the IIIabc four‐way junction in our present model was suggested (Boehringer et al, 2005; Siridechadilok et al, 2005). Because IIIb interacts with the eIF3 initiation factor complex (Sizova et al, 1998; Ji et al, 2004; Otto & Puglisi, 2004), our model places this major eIF3 binding site further away from the ribosome in excellent agreement with the direct visualization of eIF3 on the related 40S‐bound CSFV IRES with a deletion of domain II (Hashem et al, 2013).
Very recently, an independent cryo‐EM structure of a binary 80S•HCV IRES complex has been reported from the Ban laboratory at similar resolution (Quade et al, 2015). There are some differences in the number of nucleotides modeled at the 5′‐end, the apical end of domain IIIb, and the mRNA part, where the local resolution in general is lower due to flexibility. However, for the ordered parts of the IRES, a RMSD value of 2.0 Å shows that both structures are very similar.
Positioning of the mRNA part of the HCV IRES
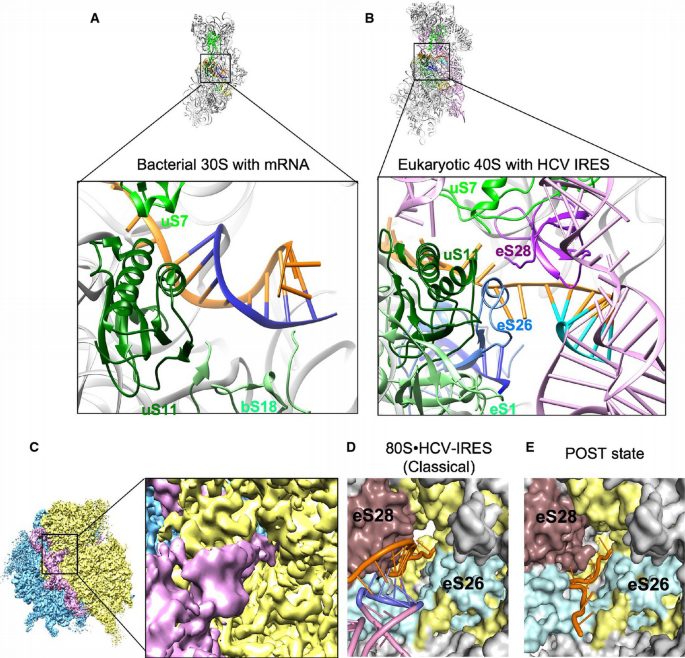
During internal initiation, the mRNA part has to be properly loaded into the mRNA binding cleft of the 40S subunit and the AUG start codon has to be positioned into the P‐site region of the 40S subunit to allow binding of the initiator tRNA. The AUG start codon is part of the domain IV stem loop. However, in the ribosome‐bound form, domain IV is melted, in agreement with findings that unwinding domain IV is necessary for function (Wang et al, 1995; Honda et al, 1996; Filbin & Kieft, 2011) and that the single‐stranded domain IV has the same environment as the E‐site region of canonical mRNA (Pisarev et al, 2008). In line with a previous proposal (Berry et al, 2010, 2011), the central domain has the important task to direct the melted domain IV via the pseudoknot helix S2 through the mRNA exit channel into the 40S mRNA binding cleft. Guiding the mRNA‐like sequence into the mRNA exit tunnel via complementary base pairing is reminiscent of bacterial translation initiation, where the Shine‐Dalgarno–anti‐Shine‐Dalgarno (SD–anti‐SD) mechanism results in a helical structure between the SD sequence upstream of the mRNA start codon and the 3′ end of the 16S rRNA. Comparison shows a similar position of both helices, thus suggesting a SD–anti‐SD mimicry during HCV IRES‐driven initiation (Fig 6A and B). However, here the anti‐SD‐like sequence is not provided in trans by 18S rRNA but in cis by the IIIf loop of the ribosome‐bound HCV IRES. This is in line with the functional importance of the pseudoknot, as mutations destroying the base pairing of helix S2 severely impair IRES activity (Berry et al, 2010). This interaction is further stabilized by eS28 (Fig 6C and D). This interaction is facilitated by an unusual path of the HCV IRES mRNA, which is different from the path of canonical mRNA in the elongating POST state (Behrmann et al, 2015) (Fig 6E).
Figure 6
SD–anti‐SD interaction in bacteria and SD–anti‐SD‐like interaction of HCV IRES at the eukaryotic mRNA exit channel
A, B Comparative solvent side views of the small subunit with the bacterial SD chamber on the 30S from the 70S POST state (PDB 2HGR) (Yusupova et al, 2006) and the corresponding region of the eukaryotic 40S in the classical 80S•HCV IRES complex. Analogous proteins and rRNA helices are highlighted in the same colors, respectively. (A) Crystal structure of the 30S subunit with mRNA. The SD helix is established between the mRNA (orange) and the 3′ end of the 16S rRNA (blue). (B) Atomic model of the 40S subunit with the HCV IRES model (pink). The mRNA part and S2 of the IRES are shown in orange and cyan, and the 3′ end of the 18S rRNA is shown in blue.
C Overview and zoomed view of classical 80S•HCV IRES. The cryo‐EM map of the classical 80S•HCV IRES was filtered according to resolution.
D, E mRNA exit channel in classical 80S•HCV IRES and POST state (PDB 5AJ0) (Behrmann et al, 2015). The two structures are aligned at the 40S body. We note that eS28 as part of the 40S head is differently located because of the head tilt. The surface representation was created using the PDB models. Shown are the 18S rRNA (yellow), ribosomal proteins (gray), eS26 (cyan), eS28 (purple), and HCV IRES (pink).
Our high‐resolution map also corroborates the proposed involvement of HCV IRES domain II in mRNA loading (Spahn et al, 2001). Domain II emerges as a protrusion from the central IRES core and arcs into the decoding cleft, where the tip of the apical loop of domain II with G82C83 is seen to contact the mRNA‐like part in the E‐site region (Figs 5B and 7A and B). This is consistent with the proposal that the domain IIb stem loop stabilizes the mRNA‐like part in the 40S mRNA binding cleft (Filbin & Kieft, 2011).
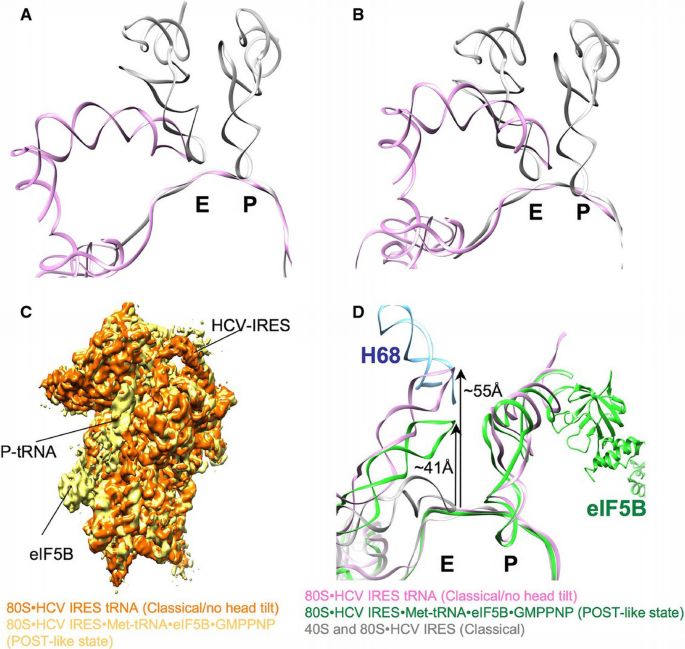
Figure 7
The apical loop of the HCV IRES domain II changes the position at the E‐site during initiation
A. Comparison of HCV IRES from 80S•HCV IRES classical (pink) and tRNAs from POST state (gray) (PDB 5AJ0) (Behrmann et al, 2015) in a common 40S body alignment of the cryo‐EM maps.
B. Same as above but in a common 40S head alignment.
C. Comparison of the 40S subunit and bound ligands of the classical 80S•HCV IRES complex (orange) and 80S•HCV IRES•Met‐tRNA•eIF5B•GMPPNP complex (POST‐like state; yellow; EMD 2683) (Yamamoto et al, 2014) in common 60S alignment.
D. The models of the 80S•HCV IRES classical complex (gray), 80S•HCV IRES•Met‐tRNA•eIF5B•GMPPNP complex (POST‐like state; green; PDB 4UPZ) (Yamamoto et al, 2014), and 80S•HCV IRES classical/no head tilt complex (pink) are rigid body fitted into the corresponding densities in a common 40S body alignment. The helix 68 from the 28S rRNA is depicted as well (blue).
Active manipulation of the 40S conformation by HCV IRES domain II
Interestingly, there are striking similarities upon ribosomal binding of the HCV IRES (Spahn et al, 2001; Filbin et al, 2013), the CrPV IRES (Schuler et al, 2006), or canonical initiation factors eIF1 and eIF1A (Passmore et al, 2007) as all ligands lead to a similar conformational change of the head of the 40S. Hence, IRESs represent built‐in mRNA translation factors (Spahn et al, 2004). This similarity between unrelated IRESs and canonical initiation may be enforced by mechanistic requirements, as all translation initiation pathways have to facilitate mRNA loading, start site selection, tRNA binding, and ribosomal subunit association. The 40S conformational change can be defined more precisely in the present cryo‐EM maps at improved resolution. In terms of overall conformation, the present major subpopulation of the binary 80S•HCV IRES complex is related to the post‐translocational (POST) state of the elongating 80S ribosome. However, the 40S head is tilted away by ~17° from the central protuberance of the 60S subunit (Fig 2A).
The same changed 40S conformation is seen in the binary 40S•HCV IRES (Fig 1E). Thus, the open conformation of the 40S subunit (Spahn et al, 2001; Filbin et al, 2013) is preserved after factor‐free ribosomal subunit association. However, in the present P‐site tRNA containing subpopulation (Figs 1D, 2A and 7C) or after eIF5B‐facilitated subunit joining in the presence of initiator tRNA (Yamamoto et al, 2014), the IRES‐induced head tilt is reversed (Fig 7C). The active manipulation of the translational apparatus by the HCV IRES depends on domain II (Spahn et al, 2001; Filbin et al, 2013), which interacts with the proteins uS7 (rpS5) and eS25 in the 40S head, as well as with uS11 (rpS14) in the platform region. The proteins uS7 and eS25 also interact with the CrPV IRES (Schuler et al, 2006; Muhs et al, 2011), and their functional importance has been demonstrated in both systems (Fukushi et al, 2001; Babaylova et al, 2009; Landry et al, 2009; Muhs et al, 2011).
In the presence of the head tilt, the apical loop of HCV IRES domain II adopts a chimeric position on the 40S subunit. From the perspective of the tilted 40S head, it overlaps with the E‐site tRNA (Filbin et al, 2013) (Fig 7B). However, from the platform perspective, it is located more beside the E‐site tRNA (Fig 7A), where it would clash with uS7 (rpS5) in the canonically oriented, untilted 40S head (Fig 5A). Thus, domain II can be regarded as a wedge that intercalates into the mRNA binding cleft and forces the 40S subunit to adopt an open conformation. Accordingly, binding of domain II to the 40S head and the closed conformation of the 40S appear mutually exclusive. Indeed, when the IRES‐induced head tilt is backed by the presence of P‐site tRNA, a large‐scale conformational change of the HCV IRES takes place leading to a repositioning of domain II around 55 Å from the 40S E‐site to the 60S E‐site at helix 68 of 28S rRNA, where usually the acceptor stem of E‐tRNA interacts (Fig 7D).
Benzimidazole is the one of the inhibitors for HCV IRES‐driven translation initiation, and the crystal structure in complex with domain IIa was solved and shows that the drug induces a pronounced conformational change in the RNA (Dibrov et al, 2012) (Boerneke et al, 2014). Interestingly, this structure (PDB 3TZR) does not fit to the corresponding domain II regions from our 80S•HCV IRES, classical, and classical/no head tilt cryo‐EM maps. This suggests that by locking domain II in an extended form (Dibrov et al, 2012; Boerneke et al, 2014), benzimidazole prevents the productive interaction with the 40S subunit to inhibit initiation.
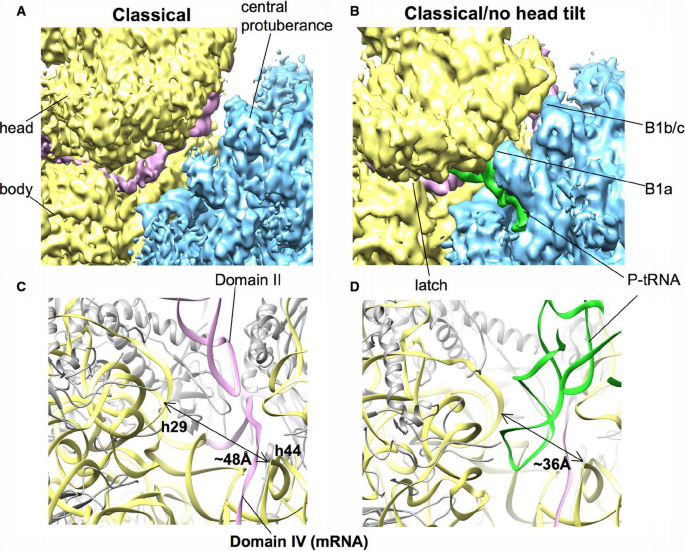
The IRES domain II induced 40S head tilt generates a gap between the central protuberance of 60S subunit and the head of 40S subunit, thereby disrupting the respective intersubunit bridges (Fig 8A and B). This in turn may explain why in the absence of initiation factors elevated concentration of Mg2+/Ca2+ ions is needed for ribosomal subunit joining (Lancaster et al, 2006). In contrast to the Dicistroviridae intergenic region IRESs (Spahn et al, 2004), the HCV IRES does not support subunit association. Moreover, the open conformation of the 40S subunit induced by domain II provides a mechanistic explanation for mRNA loading. It opens the latch of the mRNA entry tunnel (Spahn et al, 2001), which opens/widens the 40S P‐tRNA binding site from 36 Å to around 48 Å between head and platform and the mRNA exit tunnel/channel (Fig 8C and D) explaining the domain II‐dependent sensitivity of 48S complexes toward eIF1‐induced destabilization (Pestova et al, 2008). Furthermore, HCV IRES domain II prevents initiator tRNA binding in the absence of initiation factors and has been implicated in eIF5‐induced GTP hydrolysis on eIF2 leading to eIF2•GDP release from the 48S initiation complex (Locker et al, 2007; Pestova et al, 2008). It is in agreement with a recent single‐molecule FRET analysis suggesting movement of domain II in HCV IRES•40S complexes (Fuchs et al, 2015), which, however, was suppressed by the addition of cell lysate.
Figure 8
Tilt movement of the 40S head causes opening of the P‐tRNA binding site and the mRNA channel
A, B Comparison of the 80S•HCV IRES classical complex and classical/no head tilt complex (40S yellow, 60S blue, IRES pink, P‐tRNA green) from the intersubunit view. Intersubunit bridges 1b/c and 1a are indicated as B1b/c and B1a, respectively. The cryo‐EM map of classical 80S•HCV IRES classical complex was filtered to 6 Å.
C, D Close‐up of the mRNA tunnel. The distance between helix 29 and helix 44 of the 18S rRNA is indicated with an arrow and Å units.
By tuning the conformational energy landscape of the ribosome in a similar manner to protein translation factors (Munro et al, 2009), HCV IRES domain II drives translation initiation by an active mechanism (Fig 9). As domain II of the IRES does not contribute to the overall affinity of the HCV IRES to the 40S subunit (Kieft et al, 2001; Spahn et al, 2001; Filbin et al, 2013), its interaction energy may pay for an unfavorable 40S conformation. Attachment to the 40S head and opening of the 40S subunit by a wedge‐like mechanism appears to facilitate mRNA loading into the mRNA binding cleft and may improve access of the initiator tRNA. Once bound, however, closing of the 40S subunit is required to ensure tight binding of the P‐site bound tRNA for subsequent steps. The structure of the present P‐site tRNA containing subpopulation (Fig 1D) suggests that binding of tRNA to the P‐site is sufficient to restore the closed 40S conformation. Thus, there is a delicate balance and the interactions of the HCV IRES with the ribosome are tuned for the need of stable binding as well as sufficient flexibility to support internal initiation of translation in a coordinated and regulated manner.
Figure 9
The model of HCV IRES‐driven initiation factor‐independent subunit assembly
HCV IRES (pink) binds to the 40S subunit (yellow) and facilitates head tilt (orange). Elevated cations help the initiation factor‐independent 60S subunit (blue) joining. The assembled 80S•HCV IRES complex exhibits dynamic 40S subunit movement and forms classical, rolled, and rotated state, which compare to canonical translation elongation POST and PRE states. The head tilt is reversed after tRNA (green) binding to the P‐site. At this event, the tip of HCV IRES domain II is released from the mRNA region and interacts with helix 68 of 28S rRNA of 60S subunit.
Materials and Methods
Purification of components for in vitro assembly of ribosomal complexes
40S and 60S subunits from rabbit reticulocyte lysates (RRLs) were purified according to published protocols (Pisarev et al, 2007). HCV IRES (nucleotides 1–341) together with 85 nucleotides of the HCV coding sequence was transcribed and purified according to (Yamamoto et al, 2014).
Assembly of binary 80S and 40S complexes with HCV IRES
40S•HCV IRES complexes were prepared as described previously (Yamamoto et al, 2014). In order to form a 80S•HCV IRES complex, a two‐fold excess of 60S subunits was added and the mixture was incubated further for 10 min at 37°C in the presence of 2.5 mM CaCl2. The obtained 40S•HCV IRES and 80S•HCV IRES complexes were affinity purified (Yamamoto et al, 2014). For the 80S•HCV IRES complex, 2.5 mM CaCl2 was replaced by additional 2.5 mM MgCl2 (final concentration 7.5 mM) during the washing step to improve elution efficiency.
Cryo‐electron microscopy and 3D reconstruction
Aliquots of the 40S• and 80S•HCV IRES complexes were flash‐frozen in liquid ethane on carbon‐coated Quantifoil grids (Quantifoil, Germany) using a Vitrobot (FEI) device. Digital micrographs for 40S•HCV IRES complex were automatically collected on a Tecnai G2 Polara microscope (FEI) at 300 kV equipped with a F416 CMOS camera (TVIPS) using Leginon (Suloway et al, 2005). Data were collected at a defocus range between −2 and −4 μm and at a nominal magnification of 115,000× resulting in a pixel size of 0.79 Å.
Digital micrographs for 80S•HCV IRES complex were automatically collected on a Titan Krios microscope at 300 kV equipped with a Falcon II DED detector using EPU software (FEI). Data were collected at defocus range between −1 and −4 μm and at a nominal magnification of 75,000× resulting in a pixel size of 1.07 Å.
Defocus values were determined with CTFFIND3 (Mindell & Grigorieff, 2003). Particle images were preselected using Signature (Chen & Grigorieff, 2007) and screened semiautomatically. Particle images were subjected to multiparticle refinement in SPIDER (Frank et al, 1996; Loerke et al, 2010; Ratje et al, 2010).
A total of 435,801 particle images of the 80S•HCV IRES complex from 7707 images were subjected to the refinement with a low‐resolution reconstruction of a vacant rabbit 80S ribosome as starting references. After initial rounds of refinement, 88% of the images corresponding to the fully formed complex were isolated. The remaining particle images split into structures corresponding to rolled (18%) and rotated (13%) states, and 60S with an undefined 40S subunit (8%). The largest subset (49%), which relates to the POST state, was further separated into classical (39%) and classical/no head tilt (10%) using focused reassignment to the IRES density (Penczek et al, 2006). A final data set of 171,820 particle images with a pixel size 1.07 Å was selected and refined to high resolution using SPARX (Hohn et al, 2007). The final resolution of the HCV IRES•80S complex is 3.9 Å as estimated with the FSC 0.143 criterion (Fig EV1).
A total of 218,805 particle images of the 40S•HCV IRES complex from 5,692 images were subjected to refinement with a low‐resolution reconstruction of a vacant rabbit 40S ribosome as starting references. The largest, IRESs containing subsets were isolated and further analyzed. A final data set of 145,286 particle images (66%) with a pixel size 1.56 Å was selected and refined to high resolution. The final resolution of the HCV IRES‐40S complex is 6.7 Å as estimated with the FSC 0.143 criterion (Fig EV1).
Modeling of HCV IRES, interacting 8 ribosomal protein and helix 26
Docking of available atomic models of ribosomal components and available HCV IRES subdomains [Domain II (1P5P) (Lukavsky _et al_, 2003), pseudoknot domain (3T4B) (Berry et al, 2011), domain IIId (1FQZ)] (Klinck et al, 2000), CUUG tetra loop, and the adenine bulge (3U5H) (Ben‐Shem et al, 2011) was done using UCSF CHIMERA (Pettersen et al, 2004). The model of the 4‐way junction (1KH6) (Kieft et al, 2002) was flexible docked into the experimental map using MDFit (Whitford et al, 2011). Head and body of the 40S subunit and L1 stalk as well as the remaining parts of the 60S subunit (PDB 5AJ0; Behrmann et al, 2015) were fitted separately.
The real‐space refinement using Phenix was done for ribosomal proteins into the experimental map to obtain initial models (Adams et al, 2010). Poorly fitting parts were manually fitted in coot (Emsley et al, 2010) and energy minimized with REFMAC5 (Murshudov et al, 2011). Atomic models for missing parts of the HCV IRES were generated de novo or via homology modeling and connected using COOT and RCrane (Keating & Pyle, 2012) according to experimental map. Finally, ribosomal proteins and HCV IRES were structure idealized using REFMAC5. The obtained models for HCV IRES and the 8 interacting proteins were then refined together with the remaining 40S subunit using Phenix (Adams et al, 2010) (Table EV3).
Coordinates
The electron density maps of the ribosomal 80S•HCV IRES and 40S•HCV IRES complexes were deposited in the Electron Microscopy Data Bank (European Molecular Biology Laboratory‐European Bioinformatics Institute, Cambridge, UK) with the accession numbers EMD‐3221 (classical), EMD‐3223 (classical, no head tilt), EMD‐3225 (Rolled), EMD‐3226 (Rotated), and EMD‐3224 (40S•HCV IRES). Model for the 40S•HCV IRES RNA has been deposited in the Protein Data Bank database with PDB ID 5FLX.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse‐Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221
Google Scholar - Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R (2013) Structures of the human and Drosophila 80S ribosome. Nature 497: 80–85
Google Scholar - Asnani M, Kumar P, Hellen CU (2015) Widespread distribution and structural diversity of Type IV IRESs in members of Picornaviridae. Virology 478: 61–74
Google Scholar - Babaylova E, Graifer D, Malygin A, Stahl J, Shatsky I, Karpova G (2009) Positioning of subdomain IIId and apical loop of domain II of the hepatitis C IRES on the human 40S ribosome. Nucleic Acids Res 37: 1141–1151
Google Scholar - Behrmann E, Loerke J, Budkevich TV, Yamamoto K, Schmidt A, Penczek PA, Vos MR, Burger J, Mielke T, Scheerer P, Spahn CM (2015) Structural snapshots of actively translating human ribosomes. Cell 161: 845–857
Google Scholar - Ben‐Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M (2011) The structure of the eukaryotic ribosome at 3.0 A resolution. Science 334: 1524–1529
Google Scholar - Berry KE, Waghray S, Doudna JA (2010) The HCV IRES pseudoknot positions the initiation codon on the 40S ribosomal subunit. RNA 16: 1559–1569
Google Scholar - Berry KE, Waghray S, Mortimer SA, Bai Y, Doudna JA (2011) Crystal structure of the HCV IRES central domain reveals strategy for start‐codon positioning. Structure 19: 1456–1466
Google Scholar - Bhat P, Shwetha S, Sharma DK, Joseph AP, Srinivasan N, Das S (2015) The beta hairpin structure within ribosomal protein S5 mediates interplay between domains II and IV and regulates HCV IRES function. Nucleic Acids Res 43: 2888–2901
Google Scholar - Boehringer D, Thermann R, Ostareck‐Lederer A, Lewis JD, Stark H (2005) Structure of the hepatitis C virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure 13: 1695–1706
Google Scholar - Boerneke MA, Dibrov SM, Gu J, Wyles DL, Hermann T (2014) Functional conservation despite structural divergence in ligand‐responsive RNA switches. Proc Natl Acad Sci USA 111: 15952–15957
Google Scholar - Budkevich T, Giesebrecht J, Altman RB, Munro JB, Mielke T, Nierhaus KH, Blanchard SC, Spahn CM (2011) Structure and dynamics of the mammalian ribosomal pretranslocation complex. Mol Cell 44: 214–224
Google Scholar - Budkevich TV, Giesebrecht J, Behrmann E, Loerke J, Ramrath DJ, Mielke T, Ismer J, Hildebrand PW, Tung CS, Nierhaus KH, Sanbonmatsu KY, Spahn CM (2014) Regulation of the mammalian elongation cycle by subunit rolling: a eukaryotic‐specific ribosome rearrangement. Cell 158: 121–131
Google Scholar - Cate JH, Yusupov MM, Yusupova GZ, Earnest TN, Noller HF (1999) X‐ray crystal structures of 70S ribosome functional complexes. Science 285: 2095–2104
Google Scholar - Chen JZ, Grigorieff N (2007) SIGNATURE: a single‐particle selection system for molecular electron microscopy. J Struct Biol 157: 168–173
Google Scholar - Devaraj A, Shoji S, Holbrook ED, Fredrick K (2009) A role for the 30S subunit E site in maintenance of the translational reading frame. RNA 15: 255–265
Google Scholar - Dibrov SM, Ding K, Brunn ND, Parker MA, Bergdahl BM, Wyles DL, Hermann T (2012) Structure of a hepatitis C virus RNA domain in complex with a translation inhibitor reveals a binding mode reminiscent of riboswitches. Proc Natl Acad Sci USA 109: 5223–5228
Google Scholar - Dibrov SM, Parsons J, Carnevali M, Zhou S, Rynearson KD, Ding K, Garcia Sega E, Brunn ND, Boerneke MA, Castaldi MP, Hermann T (2014) Hepatitis C virus translation inhibitors targeting the internal ribosomal entry site. J Med Chem 57: 1694–1707
Google Scholar - Easton LE, Locker N, Lukavsky PJ (2009) Conserved functional domains and a novel tertiary interaction near the pseudoknot drive translational activity of hepatitis C virus and hepatitis C virus‐like internal ribosome entry sites. Nucleic Acids Res 37: 5537–5549
Google Scholar - Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501
Google Scholar - Filbin ME, Kieft JS (2011) HCV IRES domain IIb affects the configuration of coding RNA in the 40S subunit's decoding groove. RNA 17: 1258–1273
Google Scholar - Filbin ME, Vollmar BS, Shi D, Gonen T, Kieft JS (2013) HCV IRES manipulates the ribosome to promote the switch from translation initiation to elongation. Nat Struct Mol Biol 20: 150–158
Google Scholar - Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj Leith A (1996) SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol 116: 190–199
Google Scholar - Fuchs G, Petrov AN, Marceau CD, Popov LM, Chen J, O'Leary SE, Wang R, Carette JE, Sarnow P, Puglisi JD (2015) Kinetic pathway of 40S ribosomal subunit recruitment to hepatitis C virus internal ribosome entry site. Proc Natl Acad Sci USA 112: 319–325
Google Scholar - Fukushi S, Okada M, Stahl J, Kageyama T, Hoshino FB, Katayama K (2001) Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis C virus. J Biol Chem 276: 20824–20826
Google Scholar - Hajarizadeh B, Grebely J, Dore GJ (2013) Epidemiology and natural history of HCV infection. Nat Rev Gastroenterol Hepatol 10: 553–562
Google Scholar - Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Pestova TV, Hellen CU, Frank J (2013) Hepatitis‐C‐virus‐like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature 503: 539–543
Google Scholar - Hellen CU, de Breyne S (2007) A distinct group of hepacivirus/pestivirus‐like internal ribosomal entry sites in members of diverse picornavirus genera: evidence for modular exchange of functional noncoding RNA elements by recombination. J Virol 81: 5850–5863
Google Scholar - Hohn M, Tang G, Goodyear G, Baldwin PR, Huang Z, Penczek PA, Yang C, Glaeser RM, Adams PD, Ludtke SJ (2007) SPARX, a new environment for Cryo‐EM image processing. J Struct Biol 157: 47–55
Google Scholar - Honda M, Brown EA, Lemon SM (1996) Stability of a stem‐loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA 2: 955–968
Google Scholar - Jackson RJ, Hellen CU, Pestova TV (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127
Google Scholar - Ji H, Fraser CS, Yu Y, Leary J, Doudna JA (2004) Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc Natl Acad Sci USA 101: 16990–16995
Google Scholar - Joseph AP, Bhat P, Das S, Srinivasan N (2014) Re‐analysis of cryoEM data on HCV IRES bound to 40S subunit of human ribosome integrated with recent structural information suggests new contact regions between ribosomal proteins and HCV RNA. RNA Biol 11: 891–905
Google Scholar - Jubin R (2001) Hepatitis C IRES: translating translation into a therapeutic target. Curr Opin Mol Ther 3: 278–287
Google Scholar - Kalliampakou KI, Psaridi‐Linardaki L, Mavromara P (2002) Mutational analysis of the apical region of domain II of the HCV IRES. FEBS Lett 511: 79–84
Google Scholar - Keating KS, Pyle AM (2012) RCrane: semi‐automated RNA model building. Acta Crystallogr D Biol Crystallogr 68: 985–995
Google Scholar - Khatter H, Myasnikov AG, Natchiar SK, Klaholz BP (2015) Structure of the human 80S ribosome. Nature 520: 640–645
Google Scholar - Kieft JS, Zhou K, Jubin R, Murray MG, Lau JY, Doudna JA (1999) The hepatitis C virus internal ribosome entry site adopts an ion‐ dependent tertiary fold. J Mol Biol 292: 513–529
Google Scholar - Kieft JS, Zhou K, Jubin R, Doudna JA (2001) Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA 7: 194–206
Google Scholar - Kieft JS, Zhou K, Grech A, Jubin R, Doudna JA (2002) Crystal structure of an RNA tertiary domain essential to HCV IRES‐mediated translation initiation. Nat Struct Biol 9: 370–374
Google Scholar - Klinck R, Westhof E, Walker S, Afshar M, Collier A, Aboul‐Ela F (2000) A potential RNA drug target in the hepatitis C virus internal ribosomal entry site. RNA 6: 1423–1431
Google Scholar - Kolupaeva VG, Pestova TV, Hellen CU (2000) An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J Virol 74: 6242–6250
Google Scholar - Kucukelbir A, Sigworth FJ, Tagare HD (2014) Quantifying the local resolution of cryo‐EM density maps. Nat Methods 11: 63–65
Google Scholar - Laing C, Schlick T (2009) Analysis of four‐way junctions in RNA structures. J Mol Biol 390: 547–559
Google Scholar - Laletina E, Graifer D, Malygin A, Ivanov A, Shatsky I, Karpova G (2006) Proteins surrounding hairpin IIIe of the hepatitis C virus internal ribosome entry site on the human 40S ribosomal subunit. Nucleic Acids Res 34: 2027–2036
Google Scholar - Lancaster AM, Jan E, Sarnow P (2006) Initiation factor‐independent translation mediated by the hepatitis C virus internal ribosome entry site. RNA 12: 894–902
Google Scholar - Landry DM, Hertz MI, Thompson SR (2009) RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev 23: 2753–2764
Google Scholar - Locker N, Easton LE, Lukavsky PJ (2007) HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J 26: 795–805
Google Scholar - Loerke J, Giesebrecht J, Spahn CM (2010) Multiparticle cryo‐EM of ribosomes. Methods Enzymol 483: 161–177
Google Scholar - Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P, Puglisi JD (2000) Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat Struct Biol 7: 1105–1110
Google Scholar - Lukavsky PJ, Kim I, Otto GA, Puglisi JD (2003) Structure of HCV IRES domain II determined by NMR. Nat Struct Biol 10: 1033–1038
Google Scholar - Lukavsky PJ (2009) Structure and function of HCV IRES domains. Virus Res 139: 166–171
Google Scholar - Luttermann C, Meyers G (2009) The importance of inter‐ and intramolecular base pairing for translation reinitiation on a eukaryotic bicistronic mRNA. Genes Dev 23: 331–344
Google Scholar - Malygin AA, Kossinova OA, Shatsky IN, Karpova GG (2013a) HCV IRES interacts with the 18S rRNA to activate the 40S ribosome for subsequent steps of translation initiation. Nucleic Acids Res 41: 8706–8714
Google Scholar - Malygin AA, Shatsky IN, Karpova GG (2013b) Proteins of the human 40S ribosomal subunit involved in hepatitis C IRES binding as revealed from fluorescent labeling. Biochemistry 78: 53–59
Google Scholar - Matsuda D, Mauro VP (2014) Base pairing between hepatitis C virus RNA and 18S rRNA is required for IRES‐dependent translation initiation in vivo. Proc Natl Acad Sci USA 111: 15385–15389
Google Scholar - Mindell JA, Grigorieff N (2003) Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142: 334–347
Google Scholar - Muhs M, Yamamoto H, Ismer J, Takaku H, Nashimoto M, Uchiumi T, Nakashima N, Mielke T, Hildebrand PW, Nierhaus KH, Spahn CM (2011) Structural basis for the binding of IRES RNAs to the head of the ribosomal 40S subunit. Nucleic Acids Res 39: 5264–5275
Google Scholar - Munro JB, Sanbonmatsu KY, Spahn CM, Blanchard SC (2009) Navigating the ribosome's metastable energy landscape. Trends Biochem Sci 34: 390–400
Google Scholar - Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67: 355–367
Google Scholar - Odreman‐Macchioli F, Baralle FE, Buratti E (2001) Mutational analysis of the different bulge regions of hepatitis C virus domain II and their influence on internal ribosome entry site translational ability. J Biol Chem 276: 41648–41655
Google Scholar - Otto GA, Lukavsky PJ, Lancaster AM, Sarnow P, Puglisi JD (2002) Ribosomal proteins mediate the hepatitis C virus IRES‐HeLa 40S interaction. RNA 8: 913–923
Google Scholar - Otto GA, Puglisi JD (2004) The pathway of HCV IRES‐mediated translation initiation. Cell 119: 369–380
Google Scholar - Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V (2007) The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell 26: 41–50
Google Scholar - Penczek PA, Frank J, Spahn CM (2006) A method of focused classification, based on the bootstrap 3D variance analysis, and its application to EF‐G‐dependent translocation. J Struct Biol 154: 184–194
Google Scholar - Perard J, Leyrat C, Baudin F, Drouet E, Jamin M (2013) Structure of the full‐length HCV IRES in solution. Nat Commun 4: 1612
Google Scholar - Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU (1998) A prokaryotic‐like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev 12: 67–83
Google Scholar - Pestova TV, de Breyne S, Pisarev AV, Abaeva IS, Hellen CU (2008) eIF2‐dependent and eIF2‐independent modes of initiation on the CSFV IRES: a common role of domain II. EMBO J 27: 1060–1072
Google Scholar - Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612
Google Scholar - Pisarev AV, Unbehaun A, Hellen CU, Pestova TV (2007) Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol 430: 147–177
Google Scholar - Pisarev AV, Kolupaeva VG, Yusupov MM, Hellen CU, Pestova TV (2008) Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J 27: 1609–1621
Google Scholar - Quade N, Boehringer D, Leibundgut M, van den Heuvel J, Ban N (2015) Cryo‐EM structure of Hepatitis C virus IRES bound to the human ribosome at 3.9‐A resolution. Nat Commun 6: 7646
Google Scholar - Ratje AH, Loerke J, Mikolajka A, Brunner M, Hildebrand PW, Starosta AL, Donhofer A, Connell SR, Fucini P, Mielke T, Whitford PC, Onuchic JN, Yu Y, Sanbonmatsu KY, Hartmann RK, Penczek PA, Wilson DN, Spahn CM (2010) Head swivel on the ribosome facilitates translocation by means of intra‐subunit tRNA hybrid sites. Nature 468: 713–716
Google Scholar - Reynolds JE, Kaminski A, Carroll AR, Clarke BE, Rowlands DJ, Jackson RJ (1996) Internal initiation of translation of hepatitis C virus RNA: the ribosome entry site is at the authentic initiation codon. RNA 2: 867–878
Google Scholar - Schuler M, Connell SR, Lescoute A, Giesebrecht J, Dabrowski M, Schroeer B, Mielke T, Penczek PA, Westhof E, Spahn CM (2006) Structure of the ribosome‐bound cricket paralysis virus IRES RNA. Nat Struct Mol Biol 13: 1092–1096
Google Scholar - Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E (2005) Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science 310: 1513–1515
Google Scholar - Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CU (1998) Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J Virol 72: 4775–4782
Google Scholar - Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J (2001) Hepatitis C virus IRES RNA‐induced changes in the conformation of the 40s ribosomal subunit. Science 291: 1959–1962
Google Scholar - Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J (2004) Cryo‐EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA‐based translation factor. Cell 118: 465–475
Google Scholar - Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B (2005) Automated molecular microscopy: the new Leginon system. J Struct Biol 151: 41–60
Google Scholar - Terenin IM, Dmitriev SE, Andreev DE, Shatsky IN (2008) Eukaryotic translation initiation machinery can operate in a bacterial‐like mode without eIF2. Nat Struct Mol Biol 15: 836–841
Google Scholar - Visweswaraiah J, Pittman Y, Dever TE, Hinnebusch AG (2015) The beta‐hairpin of 40S exit channel protein Rps5/uS7 promotes efficient and accurate translation initiation in vivo. Elife 4: e07939
Google Scholar - Voorhees RM, Fernandez IS, Scheres SH, Hegde RS (2014) Structure of the mammalian ribosome‐Sec61 complex to 3.4 A resolution. Cell 157: 1632–1643
Google Scholar - Wang C, Le SY, Ali N, Siddiqui A (1995) An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA 1: 526–537
Google Scholar - Whitford PC, Ahmed A, Yu Y, Hennelly SP, Tama F, Spahn CM, Onuchic JN, Sanbonmatsu KY (2011) Excited states of ribosome translocation revealed through integrative molecular modeling. Proc Natl Acad Sci USA 108: 18943–18948
Google Scholar - Willcocks MM, Locker N, Gomwalk Z, Royall E, Bakhshesh M, Belsham GJ, Idamakanti N, Burroughs KD, Reddy PS, Hallenbeck PL, Roberts LO (2011) Structural features of the Seneca Valley virus internal ribosome entry site (IRES) element: a picornavirus with a pestivirus‐like IRES. J Virol 85: 4452–4461
Google Scholar - Xue S, Tian S, Fujii K, Kladwang W, Das R, Barna M (2015) RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature 517: 33–38
Google Scholar - Yamamoto H, Unbehaun A, Loerke J, Behrmann E, Collier M, Burger J, Mielke T, Spahn CM (2014) Structure of the mammalian 80S initiation complex with initiation factor 5B on HCV‐IRES RNA. Nat Struct Mol Biol 21: 721–727
Google Scholar - Yusupova G, Jenner L, Rees B, Moras D, Yusupov M (2006) Structural basis for messenger RNA movement on the ribosome. Nature 444: 391–394
Google Scholar - Zinoviev A, Hellen CU, Pestova TV (2015) Multiple mechanisms of reinitiation on bicistronic calicivirus mRNAs. Mol Cell 57: 1059–1073
Google Scholar
Acknowledgements
We thank Anett Unbehaun and Elmar Behrmann for discussion and Christine Gotthold for the preparation of ribosomes. This work was supported by grants from the Volkswagenstiftung, German‐Research Foundation (DFG; Forschergruppe 1805) to CMTS, DFG Cluster of Excellence ‘Unifying Concepts in Catalysis’ ‐ Research Field E3‐1 to PS and SFB740 to CMTS, PS, and PWH.
Author information
Author notes
- These authors contributed equally to this work
Authors and Affiliations
- Institut für Medizinische Physik und Biophysik, Charité – Universitätsmedizin, Berlin, Germany
Hiroshi Yamamoto, Marianne Collier, Justus Loerke, Jochen Ismer, Andrea Schmidt, Tarek Hilal, Thiemo Sprink, Kaori Yamamoto, Thorsten Mielke, Jörg Bürger, Marylena Dabrowski, Peter W Hildebrand, Patrick Scheerer & Christian MT Spahn - UltraStrukturNetzwerk, Max Planck Institute for Molecular Genetics, Berlin, Germany
Thorsten Mielke & Jörg Bürger - Structural Biology Programme, CEITEC, Masaryk University, Brno, Czech Republic
Tanvir R Shaikh
Authors
- Hiroshi Yamamoto
- Marianne Collier
- Justus Loerke
- Jochen Ismer
- Andrea Schmidt
- Tarek Hilal
- Thiemo Sprink
- Kaori Yamamoto
- Thorsten Mielke
- Jörg Bürger
- Tanvir R Shaikh
- Marylena Dabrowski
- Peter W Hildebrand
- Patrick Scheerer
- Christian MT Spahn
Contributions
HY, MC, MD and CMTS designed experiments. HY and MC prepared complexes. TM, JB and TRS collected cryo‐EM data. HY, MC, JL and CMTS processed images. HY, MC, JI, AS, TH, TS, PWH, PS and CMTS built the model. HY, MC and KY prepared figures. HY, MC, and CMTS discussed results and wrote the paper.
Corresponding author
Correspondence toChristian MT Spahn.
Ethics declarations
The authors declare that they have no conflict of interest.
Additional information
The EMBO Journal (2015) 34: 3042–3058
Supplementary Information
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Review Process File
EV1
Multiparticle refinement for 80S•HCV IRES complex
Multiparticle refinement was used to overcome heterogeneity of the complex. The largest population (88%) containing strong IRES density was isolated and further separated into rolled (18%) and rotated (13%) states, which are related to PRE state, and a subpopulation corresponding to a 60S subunit with an undefined 40S subunit (8%). The major subpopulation (49%), which is related to the POST state, was further separated into classical (39%) and classical/no head tilt (10%) by focused reassignment using a mask on the IRES density. Thereby, density for P‐site tRNA emerged for the classical/no head tilt subpopulation. The classical 80S•HCV IRES subpopulation was further refined to high resolution. The cryo‐EM maps were filtered according to resolution. HCV IRES (pink), 40S subunit (yellow), 60S subunit (blue), tRNA (green).
EV2
Overall fitting of the P‐tRNA derived from POST state to the density of classical/no head tilt 80S•HCV IRES complex
Isolated density from the 80S•HCV IRES classical/no head tilt complex in the P‐site region (gray, semitransparent) superposed with a model for the P‐tRNA (PDB 5AJ0) (Behrmann et al, 2015). The cryo‐EM map was filtered according to resolution.
EV3
Resolution curves of the 80S•HCV IRES classical, rolled, rotated, classical/no head tilt, and 40S•HCV IRES complexes
The final resolution of the 80S•HCV IRES classical (red), rolled (green dot), rotated (blue dot), classical/no head tilt (purple dot), and 40S•HCV IRES complex (light blue dot) is 3.9, 5.6, 5.8, 5.8 and 6.7 Å, respectively, and was estimated using a cutoff of 0.143 of the Fourier shell correlation (FSC) standard curves.
EV4
Local resolution estimation of the 80S•HCV IRES Classical complex
The local resolution was calculated with ResMap (Kucukelbir et al, 2014) and is color‐coded in Å units.
A 80S•HCV IRES classical, factor binding side view on the full cryo‐EM map (top left), vertical slice in a factor binding side view (top right), isolated HCV IRES density (bottom left). Histogram for IRES (pink), 80S or 40S (yellow), complex (green) (bottom right). The values were individually normalized since the IRES has too small values than the others due to size of density.
B‐E 80S•HCV IRES Rolled (B), Rotated (C), Classical/no head tilt (D) and 40S•HCV IRES (E).
Data information: The cryo‐EM map was filtered according to resolution.
EV5
Building a tertiary structure model of the ribosome‐bound HCV IRES RNA
Overlay of the experimental map (gray, semitransparent) and the docked structures of the isolated HCV IRES domain II (PDB code 1P5P; Lukavsky et al, 2003), pseudoknot domain (3T4B), domain IIId (PDB 1FQZ and 3U5H), adenine bulge (PDB 3U5H), 4‐way junction (PDB 1KH6), and the final models. Domains are colored according to Fig 3B. The cryo‐EM map was filtered according to resolution.
EV6
The interaction details between ribosomal proteins and HCV IRES
Close‐up view on 3 interaction sites of HCV IRES and the ribosomal proteins (eS1, eS27 and eS7) at 40S body region. Same color code as in Fig 3B.
Rights and permissions
About this article
Cite this article
Yamamoto, H., Collier, M., Loerke, J. et al. Molecular architecture of the ribosome‐bound Hepatitis C Virus internal ribosomal entry site RNA.EMBO J 34, 3042–3058 (2015). https://doi.org/10.15252/embj.201592469
- Received: 06 July 2015
- Revised: 28 October 2015
- Accepted: 29 October 2015
- Published: 24 November 2015
- Issue date: 14 December 2015
- DOI: https://doi.org/10.15252/embj.201592469