PET Imaging of CCND1 mRNA in Human MCF7 Estrogen Receptor–Positive Breast Cancer Xenografts with Oncogene-Specific [64Cu]Chelator-Peptide Nucleic Acid-IGF1 Analog Radiohybridization Probes (original) (raw)
OtherBASIC SCIENCE INVESTIGATIONS
, Mohan R. Aruva, Kaijun Zhang, Nylla Shanthly, Christopher A. Cardi, Mathew L. Thakur and Eric Wickstrom
Journal of Nuclear Medicine October 2007, 48 (10) 1699-1707; DOI: https://doi.org/10.2967/jnumed.107.042499

Abstract
Treatment of breast cancer is hampered by a large unmet need for rapid, sensitive, specific staging and stratification of palpable and nonpalpable abnormalities. Mammography and physical examination miss many early breast cancers, yet detect many benign lesions. Cyclin D1, encoded by CCND1 messenger RNA (mRNA), and insulin-like growth factor 1 receptor (IGF1R) are key regulators of cell proliferation that are overexpressed in most breast cancers. Therefore, we hypothesized that malignant breast masses could be imaged and quantitated externally by PET with a dual-specificity probe that targets both CCND1 mRNA and IGF1R. Methods: We designed a _CCND1_-specific peptide nucleic acid (PNA) hybridization sequence (CTGGTGTTCCAT), separated by a C-terminal spacer to a cyclized IGF1 peptide analog (d-Cys-Ser-Lys-Cys), for IGF1R-mediated endocytosis. On the N-terminus we attached a chelator (1,4,7-tris(carboxymethylaza)cyclododecane-10-azaacetyl [DO3A]) for the positron-emitting nuclide 64Cu. We administered the [64Cu]_CCND1_-IGF1 analog radiohybridization probes, as well as sequence controls, by tail vein to immunocompromised female NCr mice bearing human MCF7 estrogen-dependent, receptor-positive xenografts. We imaged the mice by PET and CT 4 and 24 h later, and measured tissue distribution of the radiohybridization probes. Results: We observed 8 ± 2-fold higher PET intensity in the center of the breast cancer xenografts than in the contralateral tissues at 24 h after injection of the [64Cu]_CCND1_-IGF1 analog radiohybridization probe. IGF1 blocking yielded significantly weaker images (P < 0.05) relative to the tumor-free side at 24 h after injection, as did a PNA mismatch probe, a peptide mismatch probe, and free 64CuCl2. Conclusion: These results are consistent with our hypothesis for radiohybridization PET of overexpressed CCND1 mRNA, dependent on IGF1R-mediated endocytosis, in suspect masses. Early noninvasive detection of initial cancerous transformation, as well as invasive or recurrent breast cancer, with dual-specificity radiohybridization probes, might enable molecularly targeted staging, stratification, and choice of therapy.
Cyclin D1 (BCL1, PRAD1, CCND1) is a proto-oncogenic regulator (1) of the G1/S checkpoint in the cell cycle that has been implicated in the pathogenesis of several types of cancer, including breast cancer. The cyclin D1 protein is overexpressed in ≈80% of breast cancers (2) but shows low expression in fibroadenomas (3) or normal breast tissues (4). Neoplastic transformation is often related to abnormal activation of growth factor receptors and their signaling pathways. Experimental and clinical data strongly suggest that the insulin-like growth factor (IGF1) receptor (IGF1R) is involved in breast cancer development and progression (5). The IGF1R gene is amplified in ≈70% of human tumors, particularly in metastatic cells (6). IGF1R internalizes IGF1 into endosomes that acidify, releasing cargo to the cytoplasm, followed by recycling of IGF1R back to the cell surface (7).
Direct imaging of overexpressed oncogene messenger RNA (mRNA) could provide information on cellular gene expression patterns and might reveal molecular changes in disease states at relatively early stages (8), providing opportunities for genetic therapy, especially against overexpressed oncogene mRNAs (9). Radiohybridization PET imaging of breast cancer gene expression (Fig. 1) after mammography might distinguish malignant masses from benign growths and enable lesion staging and stratification noninvasively.
Naturally occurring oligonucleotides cannot be used directly for in vivo imaging because they are rapidly degraded in vivo by endonucleases and exonucleases (9). Many oligonucleotide modifications are available that degrade more slowly (9), but peptide nucleic acids (PNAs) are completely resistant to both nucleases and proteases (11).
Due to their uncharged backbones, PNAs hybridize to RNA more strongly and specifically than most oligonucleotide derivatives (11). Twelve PNA bases are sufficient for statistical uniqueness among transcribed mRNAs, and our melting temperature results with peptide-PNA-peptides hybridized to RNA confirmed that 12 PNA residues hybridize strongly enough and specifically enough to serve as mRNA probes in vivo (12).
FIGURE 1.
Schematic of [64Cu]DO3A-PNA-IGF1 analog, designed to bind to the receptor for IGF1, internalize, and hybridize with CCND1 mRNA. PET of γ-particles created by electron annihilation of positrons from decaying 64Cu might identify sites of high CCND1 mRNA expression in breast cancer xenografts. (Adapted with permission of (10).)
The neutral PNA backbone does not show any propensity to associate nonspecifically with intracellular nucleic acid-binding proteins that normally bind negatively charged oligonucleotide analogs (11). This facilitates effective efflux of unbound PNA probes, increasing the signal-to-noise ratio for probes bound to mRNA. Unlike normal oligonucleotide or phosphorothioate analogs, PNA hybridization does not induce ribonuclease H degradation of bound mRNA (11), which would eliminate the mRNA target that we wish to image.
Mammalian cells internalize underivatized PNAs quite slowly (13). However, conjugation of basic peptides elevates PNA uptake by all cells nonspecifically (14). To target specifically cells that overexpress IGF1R, a cyclized d-peptide analog of IGF1, d(Cys-Ser-Lys-Cys), coupled to the N-terminus of an IGF1R PNA 12-mer, provided IGF1R specificity and resulted in cytoplasmic localization without sequestration in punctate vesicles (15). d-Amino acids were used to slow protease attack and were coupled in the reverse order from the normal l-amino acid sequence to recreate the original side-chain presentation (16).
Subsequently, we designed and synthesized chelator peptide-PNA-IGF1 analogs, which displayed the same hybridization strength as unmodified PNAs on binding to complementary RNA targets (12). To test the hypothesis that a complementary chelator peptide-PNA-peptide would bind strongly and specifically to an oncogene mRNA target, we measured quantitative reverse transcription-polymerase chain reaction (QRT-PCR) yields of MYC replicons on total human MCF7 estrogen receptor–positive (ER+) breast cancer cell RNA. This revealed specific blocking of the RT step by peptide-PNA-IGF1 analogs, consistent with the hypothesis that the complementary PNA probe would bind strongly to the mRNA target (17).
We tested the hypothesis that a chelator peptide-PNA-IGF1 analog would be taken up by human MCF7 ER+ estrogen-dependent metastatic breast cancer cells, which overexpress IGF1 receptor when stimulated by 17-β-estradiol (18), essential for tumorigenesis (19). QRT-PCR analysis revealed that stimulated MCF7 cells express ∼6,000 CCND1 mRNA per cell (20). As before (15), the fluorescent chelator peptide-PNA-IGF1 analog showed elevated cytoplasmic uptake in MCF7 cells, but the peptide control with 2 d-Ala residues in the middle did not (20). Planar γ-particle imaging revealed bright tumor signals of intravenously administered [99mTc]chelator peptide-PNA-IGF1 analog radiohybridization probes specific for CCND1 mRNA and MYC mRNA in human MCF7 ER+ xenografts (20,21). Control sequences with no PNA, with mismatched PNA, or with mismatched peptide yielded weak tumor signals.
Three-dimensional microPET provides better spatial and temporal resolution and sensitivity than 2-dimensional planar γ-particle scintigraphic imaging for real-time imaging in live small animals (22). Furthermore, tissue uptake can be quantitated using PET image intensities (22). 64Cu decays (β+ 653 keV, 17.4%; β− 579 keV, 39%) to 64Ni; its half-life of 12.7 h allows imaging studies to be performed for up to 24 h after administration. For reliable and reproducible 64Cu labeling, we chose a macrocyclic chelator, 1,4,7-tris(carboxymethylaza)cyclododecane-10-azaacetyl (DO3A), as a stronger chelating agent (22) than the Gly-d(Ala)-Gly-Gly used for 99mTc labeling (20).
To test our hypothesis that dual-specificity PET hybridization imaging would improve the resolution and sensitivity of cancer gene mRNA imaging, we performed microPET studies of CCND1 mRNA in human MCF7 (ER+) breast cancer xenografts.
MATERIALS AND METHODS
Design and Synthesis of DO3A-PNA-Peptide Radiohybridization Probes
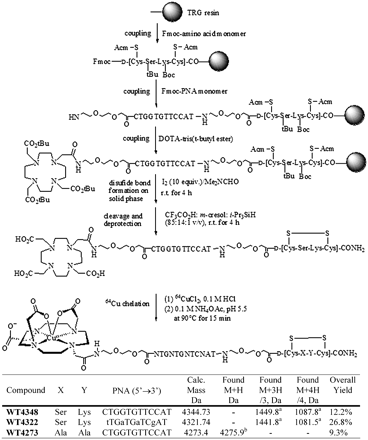
The synthesis of [64Cu]DO3A-PNA-peptide hybridization probes, with [64Cu(II)]DOTA stereochemistry based on the crystal structure (23), is illustrated in Scheme 1. Our design involved 4 aspects: (i) a receptor-specific d-peptide analog, cyclized with a Cys-Cys disulfide bridge, to enable PNA probes for cell-type specific delivery; (ii) a PNA sequence targeting the start codon region of an overexpressed oncogene mRNA, because the start codon region is usually available for PNA hybridization; (iii) a DO3A chelator that strongly binds a positron-emitting radionuclide; and (iv) hydrophilic 8-amino-3,6-dioxaoctanoic acid (aminoethoxyethoxyacetate [AEEA]) spacers at the N-terminus and at the C-terminus of the PNA hybridization sequence to minimize the steric hindrance of the bulky chelator-metal ion complex and the cyclic peptide moieties that might otherwise affect the hybridization efficiency of the PNA.
The DO3A-CCND1 PNA-IGF1 analog (WT4348) was synthesized with acceptable yields (Scheme 1) continuously on NovaSyn TentaGel Rink (TGR) resin (0.1–0.17 mmol/g; Novabiochem) on a 30-μmol scale using standard solid-phase Fmoc coupling activated by 2-(7-aza-1H-benzotriazole-1-yl)-1,1,3,3-tetramethyl-uronium-hexafluorophosphate (HATU) on an PS3 peptide synthesizer (Protein Technologies, Inc.). Two control chimeras were synthesized (Scheme 1): one (WT4322) with 4 mismatched bases that cannot hybridize to CCND1 mRNA, and another (WT4273) with 2 mismatched amino acid residues that does not enable IGF1R-mediated uptake.
The coupling times were optimized for amino acid monomer coupling (45–60 min), PNA monomer coupling (25–35 min), and tris(t_-butyl)-1,4,7,10-tetra(carboxymethylaza)cyclododecane (DOTA) coupling (60 min). Double coupling was not required. The cysteine residues were cyclized on the solid phase with 10 equivalents of I2 in (CH3)2NCHO for 4 h at room temperature (12_). Next, the resin was washed with (CH3)2NCHO and MeOH and dried under vacuum, and then cleaved and deprotected with CF3CO2H/_m_-cresol/_i_-Pr3SiH (85:14:1) for 4 h at room temperature. Cold EtOEt was added to the CF3CO2H solution to precipitate the off-white solid product.
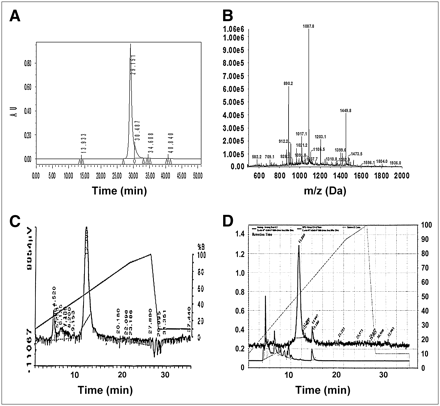
The probes were purified by reversed-phase high-performance liquid chromatography (HPLC) on a Microsorb C18 column (300-Å pore, 10-μm particle size, 10 × 250 mm; Varian, Inc.) eluted with a linear gradient of 0%–60% CH3CN in aqueous 0.1% CF3CO2H over 35 min at 50°C at a flow rate of 1 mL/min, monitored by absorbance at 260 nm (Fig. 2A). The major peaks were collected and then concentrated to dryness in vacuum. Products were characterized by electrospray ionization mass spectrometry (ESI/MS) on a SciEx API 3000 liquid chromatography/tandem mass spectrometry system (Applied Biosystems) (WT4348 and WT4322) (Fig. 2B).
FIGURE 2.
Characterization of cyclized CCND1 hybridization probe WT4348. (A) Preparative HPLC on 10 × 250 mm Microsorb C18 column, eluted with a 35-min gradient from 0% to 60% CH3CN in aqueous 0.1% CF3CO2H, at 1 mL/min, λ = 260 nm, at 50°C. (B) ESI mass spectrum. Calculated exact mass: 4,344.32 Da; Experimental mass: 1,449.8 Da (M +3)/3; 1,087.8 Da (M +4)/4. (C) Analytic HPLC of [64Cu]WT4348 on 4.5 × 250 mm Dynamax C18 column eluted with a 28-min gradient from 10% to 90% CH3CN in aqueous 0.1% CF3CO2H at 1 mL/min, at 25°C. Percent CH3CN is shown on right _y_-axis, and radiometric γ-particle emission is shown on the _y_-axis. (D) Analytic HPLC of [64Cu]WT4348 recovered from urine of tumor-bearing mice 2 h after administration, chromatographed as in C.
Cell Lines and Animal Model
Human MCF7 ER+ estrogen-dependent breast cancer cells that overexpress IGF1R (18) were maintained in Dulbecco's modified Eagle medium plus 5% calf serum, 50 U/mL penicillin, 5 μg/mL streptomycin, 2 mM glutamine, and 7.5 nM 17-β-estradiol (Sigma Chemical) at 37°C in a humidified incubator containing 5% CO2/95% air and cultured to 80% confluence before splitting or implantation. For tumor induction, 5–6 × 106 cells in 0.2 mL of culture medium were implanted intramuscularly through a sterile 27-gauge needle into the thighs of 6- to 8-wk-old female NCr immunocompromised mice (National Cancer Institute) and lightly anesthetized with a mixture of ketamine (200 mg/kg), xylazine (10 mg/kg), and acetopromazine (2 mg/kg) at a dose of 160 μL/25 g, as described earlier (20).
Tumors were allowed to grow to <1 cm in diameter, with mass ranges from 300 to 500 mg. Each injection included 10 mg of Matrigel (Becton-Dickinson). A 60-d release pellet containing 4.5 mg of 17-β-estradiol (Innovative Research of America) was implanted subdermally in each mouse. All animal studies were conducted in accordance with federal and state guidelines governing the laboratory use of animals, and under protocols approved by the Institutional Animal Care and Use Committee at Thomas Jefferson University. Animals were euthanized in a halothane chamber, consistent with U.S. Department of Agriculture regulations and American Veterinary Medical Association recommendations.
Radiolabeling of DO3A-PNA-Peptide Hybridization Probes
To a solution of 20 μg (∼4.5 nmol) DO3A-PNA-peptides in 20 μL of sterile water, 2 μL of 64CuCl2 (7.4–11.1 MBq [200–300 μCi], ∼1 nmol) (Washington University, St. Louis, MO, or MDS Nordion) in 0.1 M HCl and 200 μL of 0.1 M NH4OAc, pH 5.5, were added and then incubated at 90°C for 15 min (Scheme 1). The radiochemical purity of the [64Cu]DO3A-PNA-peptides was monitored by reverse-phase HPLC on a Dynamax C18 column (300-Å pore, 5-μm particle size, 4.5 × 250 mm; Varian, Inc.) eluted with aqueous 0.1% CF3CO2H in a gradient from 10% to 100% CH3CN over 28 min, at 1 mL/min, at 25°C.
The thermodynamic stability of [64Cu]DO3A-PNA-peptides after exposure to 100-fold molar excesses of diethylenetriaminepentaacetic acid (DTPA), human serum albumin, or cysteine at 22°C for 30 min was determined by reverse-phase HPLC as above. The metabolic stability of the CCND1 probe [64Cu]WT4348 was tested by administering 13 MBq (350 μCi) with a sterile 27-gauge needle into the lateral tail vein of a female NCr mouse. The mouse was euthanized 3 min later and exsanguinated; 0.5 mL of blood was sedimented for 10 min at 3,000_g_ in a heparinized polypropylene vial. Thirty microliters of the serum supernatant, containing 185 kBq (5 μCi) of 64Cu, were mixed with sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, heated, and analyzed on an 18% polyacrylamide Tris-glycine gel (Invitrogen) as previously described (20). Duplicate gels were autoradiographed or stained with Coomassie blue.
[64Cu]DO3A-PNA-Peptide Radiohybridization Probe Administration
Approximately 4 μg (1 nmol) of [64Cu]DO3A-PNA-peptide, containing 3.7–7.4 MBq (100–200 μCi), were injected with a sterile 27-gauge needle into the lateral tail veins of female NCr immunocompromised mice (n = 5) bearing human MCF7 ER+ breast cancer xenografts. To control for non–IGF1R-specific uptake of CCND1 probe [64Cu]WT4348, 50 μg of human recombinant IGF1 (PeproTech) were administered by tail vein 60 min before administration of [64Cu]WT4348 (n = 5). To control for nonspecific uptake of free 64Cu(II), 3.3 MBq (90 μCi) of 64CuCl2 were administered by tail vein (n = 5).
microPET/CT of Radiohybridization Probe Accumulation
To allow adequate time for probe distribution, permeation, endocytosis, trafficking, hybridization, and efflux of unbound probe, anesthetized tumor-bearing mice were imaged 4 and 24 h after radiohybridization probe injection, in parallel with the imaging times chosen for the previous scintigraphic imaging study (20). The mice were imaged in a MOSAIC small-animal PET scanner (Philips) (24) at 2.2-mm resolution. Mice were then imaged in a coregistered MicroCAT II small-animal CT scanner (ImTek) at 200-μm resolution. During imaging, the mice were restrained with devices designed specifically to provide a minimum of discomfort to the animal.
Up to 30 million PET counts were collected. Images were reconstructed using a fully 3-dimensional (3D) iterative reconstruction algorithm, giving a pixel size of 1 mm (25). Region-of-interest (ROI) analysis was performed digitally on each image from the tumor-bearing right flank to the tumor-free left flank, providing quantitation of PET intensities in 1 × 1 × 1 mm3 voxels across each subject. The voxel intensity maps may be compared with autoradiograms of 1-mm slices of tumor, although they include some scatter or random signals from outside the slice, and the spatial resolution of 1 mm is much lower than that of autoradiography.
Regions delineating the tumor were drawn at 50% of the maximum voxel intensity on every transverse slice that showed tumor intensity. Regions defining contralateral tissue were drawn on up to 5 transverse slices. Ratios of maximum central tumor image counting rates to contralateral tissue average counting rates were then calculated, enabling mouse-to-mouse comparisons. Statistical analysis of differences among groups was determined by applying the SEM, Kruskal–Wallis 1-way ANOVA on ranks, or the Holm–Sidak pairwise multiple-comparison procedure, using SigmaStat 3.0 (SPSS, Inc.).
Radiohybridization Probe Tissue Distribution
To assess tissue distribution, 0.74–0.925 MBq (20–25 μCi) of the [64Cu]DO3A-PNA-peptides in 0.2 mL of sterile saline were administered to female NCr mice bearing human MCF7 ER+ breast cancer xenografts (n = 5) through a lateral tail vein using a sterile 27-gauge needle for the 4- and 24-h distribution studies. Mice were then euthanized, and tissues were dissected, washed free of blood, blotted dry, weighed, and counted in an Auto-γ-5000 Spectrometer (Packard Instruments), together with a standard radioactive solution of a known quantity prepared at the time of injection. Results are expressed as percentage of injected dose per gram of tissue (%ID/g). Statistical analysis of differences among groups was determined by applying the SEM and Student t test.
RESULTS
Analysis and Yields of DO3A-PNA-Peptide Radiohybridization Probes
DOTA-tris(_t_-butyl) ester (Scheme 1) has strong steric hindrance, so that 1 h of coupling was needed to achieve high coupling yields, as indicated by the ninhydrin test for noncoupled amines. During the cleavage and deprotection steps, carbocations such as _t-_Bu+ can irreversibly alkylate disulfide bonds. To capture the small amount of the _t-_Bu+ without damaging the disulfide bridge of the chimeras, 1% _i_-Pr3SiH was included in the cleavage cocktail. Cleavage for 2 h at 25°C was not sufficient to remove all _t_-Bu groups as ESI/MS indicated (data not shown). When the cleavage time was increased to 4 h, ESI/MS (Fig. 2B) did not detect [M + _t_-Bu] or [M + 2 _t_-Bu] peaks, and preparative reverse-phase HPLC showed a strong product peak (Fig. 2A).
[64Cu]DO3A-PNA-Peptide Radiohybridization Probes
The probes were labeled with 64Cu (Scheme 1) similarly to our published procedure (26), at pH 5.5 without any reducing agents, avoiding the possibility of disulfide bond opening by reducing agents under basic conditions. 64Cu labeling yields were 95%–99%, measured by analytic HPLC (Fig. 2C) as described, with the [64Cu]probe peak eluting at 12.0 min.
[64Cu]DO3A-PNA-peptides were determined by HPLC to be thermodynamically stable to 100-fold molar excesses of DTPA, human serum albumin, or cysteine at 22°C for 30 min. The CCND1 probe [64Cu]WT4348 was administered to a mouse to test for probe stability in circulating blood. Serum prepared from a blood draw 3 min after administration was analyzed by SDS-PAGE and autoradiography. Negligible 64Cu radioactivity was observed over the mass range of 6–50 kDa. In particular, no 64Cu radioactivity was detected at 30 kDa, the mass of Cu/Zn superoxide dismutase, which is stable under denaturing conditions on SDS-PAGE (27).
Radiohybridization Probe Urinalysis
Stability of radiohybridization probes to metabolic breakdown was assessed by collecting urine from female NCr immunocompromised mice bearing human MCF7 ER+ breast cancer xenografts 2 h after injection of the CCND1 probe [64Cu]WT4348. Analytic HPLC of radioactivity in the combined deproteinized urine (Fig. 2D) revealed a small void volume peak of free 64Cu, 5% of radioactivity, and an intact probe peak eluting at 12.0 min, the same retention time as the original labeled probe (Fig. 2C), with 95% of the radioactivity. This result further indicates the stability of the agent in vivo, both the PNA segment and the d-peptide segment. Breakdown fragments were not detected, consistent with the model that the [64Cu]DO3A-PNA-IGF1 analog is resistant to proteases and nucleases in serum at 37°C (11). These results are consistent with our previous report (28) that [99mTc]peptide-PNA-peptides were recovered intact from urine, without apparent degradation of PNA or d-peptide segments.
microPET/CT Radiohybridization Probe Images
The CCND1 probe [64Cu]WT4348 yielded strong images at 4 and 24 h (Fig. 3). Free 64CuCl2, the PNA mismatch control [64Cu]WT4322, and the peptide mismatch control [64Cu]WT4273 yielded weak images. Preblocking IGF1R with recombinant human IGF1 30 min before administration of [64Cu]WT4348 reduced PET image intensities to the level of the controls.
FIGURE 3.
Transverse microPET and microCT images of immunocompromised NCr mice bearing human MCF7 ER+ xenografts on their right flanks recorded at 4 and 24 h after tail vein injection of 3.7–7.4 MBq (100–200 μCi) of CCND1 [64Cu]WT4348 hybridization probe, [64Cu]WT4273 peptide mismatch probe, [64Cu]WT4322 PNA mismatch probe, or free 64CuCl2 as an unchelated control. Yellow line on coronal CT image shows the level of transverse images. All images are displayed on the same scale. Color scale of the images was normalized to the maximum/minimum of frame to show dynamic range of tumor uptake. Ratios of maximum tumor PET image intensities to contralateral tissue average intensities were calculated for equal-sized ROI.
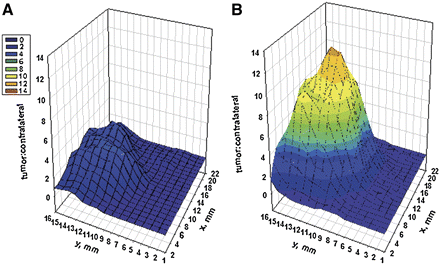
PET image intensities across serial 1-mm slices of the tumors revealed strong peaks of CCND1 probe [64Cu]WT4348 intensity in the centers of the tumors (Fig. 4). Each voxel was smaller than the tumor being imaged, so partial-volume effects were slight. The calculated tumor center/contralateral PET intensity ratios of the CCND1 probe [64Cu]WT4348 were 6.83 ± 3.41 at 4 h and 7.87 ± 1.99 at 24 h after injection.
FIGURE 4.
Ratios of tumor PET pixel counts in each 1 × 1 × 1 mm3 voxel across a central 1-mm slice of human MCF7 ER+ xenograft, vs. contralateral tissue PET counts, 4 h after administration of [64Cu]DOTA-PNA-peptide radiohybridization probes. (A) PNA mismatch probe [64Cu]WT4322 or (B) CCND1 probe [64Cu]WT4348.
Kruskal–Wallis 1-way analysis of variables determined that the 6 groups (3 probes × 2 times) were significantly different from each other (P = 0.017). When comparing each group against another by the Holm–Sidak all-pairwise multiple-comparison procedure, the CCND1 probe [64Cu]WT4348 group was significantly different from the PNA mismatch control [64Cu]WT4322 group (P = 0.011 at 4 h, and P = 0.019 at 24 h after injection) and from the peptide mismatch [64Cu]WT4273 group (P = 0.019 at 4 h, and P = 0.044 at 24 h after injection). However, the PNA mismatch group was not significantly different from the peptide mismatch group (P = 0.725 at 4 h, and P = 0.707 at 24 h after injection).
Radiohybridization Probe Tissue Distribution
The tissue distribution data of [64Cu]WT4322, [64Cu]WT4372, [64Cu]WT4348, and free 64CuCl2 in blood, tissues, and tumors are presented as %ID/g at 4 and 24 h after injection in Table 1. Both [64Cu]WT4322 and [64Cu]WT4273 control probes exhibited high kidney, liver, lung, spleen, and intestine uptake. At 24 h after injection, the CCND1 probe [64Cu]WT4348 exhibited significant liver, lung, spleen, intestine, blood, and muscle clearance, but long retention in the kidney. Within 4 h after administration, base-mismatched [64Cu]WT4322 showed significantly lower uptake compared with peptide-mismatched [64Cu]WT4273 and CCND1 probe [64Cu]WT4348, except in the kidney, which took up more [64Cu]WT4322. Both [64Cu]WT4322 and [64Cu]WT4273 showed slow clearance in kidney, liver, lung, spleen, and intestine. [64Cu]WT4348 showed the highest tumor uptake, 2.01 ± 0.43 %ID/g, and highest tumor-to-muscle ratio, 2.72 ± 0.67, at 4 h after injection. [64Cu]WT4348 also displayed the highest washout, compared with [64Cu]WT4273 and [64Cu]WT4322 (Table 1). Free 64CuCl2 accumulated preferentially over [64Cu]DO3A-PNA-peptide in most tissues. Free 64CuCl2 accumulated indistinguishably from [64Cu]DO3A-PNA-peptide in the spleen and tumor.
TABLE 1
Tissue Distribution (%ID/g) of [64Cu]DO3A-PNA-Peptide Probes
Tissue distribution of the CCND1 probe [64Cu]WT4348 versus that of the peptide mismatch [64Cu]WT4273, and [64Cu]WT4348 versus the PNA mismatch [64Cu]WT4322, were significantly different (P < 0.05) in many cases. However, at 4 h, the %ID/g of [64Cu]WT4348 versus the %ID/g of [64Cu]WT4273 was not significantly different for all 9 tissues examined. Yet at 4 h, the %ID/g of [64Cu]WT4348 versus the %ID/g of [64Cu]WT4322 was indeed significantly different (P < 0.05). At 24 h, only the kidney distributions were not significantly different for [64Cu]WT4348 versus [64Cu]WT4273 or for [64Cu]WT4348 versus [64Cu]WT4322.
DISCUSSION
There is a compelling need for early and accurate detection and stratification of malignant breast cancer. Genomic mutations lead to overexpression of the multiple genes required for malignant cell proliferation. In breast cancer cells, overexpression of CCND1 mRNA (2) and IGF1R (5) provides a clear indication of malignancy. We hypothesized that a noninvasive, dual-specificity radiohybridization imaging agent could reveal overexpression of those particular oncogenes in malignant breast cells.
Oncogene mRNA PET Images
In human MCF7 ER+ breast cancer xenografts in female immunocompromised mice, the CCND1 probe [64Cu]WT4348 displayed 7.87 ± 1.99-fold higher PET intensity in the center of the tumors than that in the contralateral tissues at 24 h after injection. Tumor contrast with [64Cu]WT4348 was significantly weaker (P < 0.05) after preadministration of an IGF1R-blocking peptide, recombinant human IGF1. A 3D plot of PET ratios of [64Cu]WT4348, across a 1-mm central slice of the tumor, serving as a low-resolution autoradiogram, demonstrated less uptake in the periphery and strong intensity in the center of the tumor. This observation suggests that IGF1R expression and CCND1 mRNA levels are highest in the centers of the MCF7 ER+ xenografts.
These results are consistent with our hypothesis that a receptor-specific targeting peptide can facilitate endocytosis of a PNA hybridization probe into the cytoplasm of cancer cells and enable external imaging of oncogene mRNA expression in vivo. Furthermore, radiohybridization imaging provided quantitation of oncogene expression levels in tumors, which can readily be translated to patients, on whom autoradiography cannot be performed. However, with a nonspecific Lys4 tail that enabled universal cell permeation, [64Cu]PNA probes of UNR mRNA in human MCF7 xenografts in another laboratory did not yield sequence-specific tumor images (29).
Radiohybridization Probe Distribution
Tissue distributions differed among the 3 probes. IGF1 blocking, however, eliminated preferential tumor accumulation. The %ID/g was measured in entire tumors after dissection, whereas the PET images revealed hot spots of CCND1 mRNA in the centers of the tumors. Therefore, the %ID/g for the entire dissected tumor does not necessarily correlate with the millimeter-by-millimeter PET voxel intensities. Active cells in the center of a small tumor are not unprecedented. In general, vascularized tumors < 1 cm metabolize strongly in their centers, with no more than 2% necrotic cells (30), unlike large tumors with necrotic cores. Even a nonspecific near-infrared fluorescent nanoparticle displayed patchy concentration in the centers of small gliosarcoma or colon adenocarcinoma xenografts (31), suggesting that vascularization is the principal driver for midtumor imaging intensity. Similarly, optical imaging of flavoprotein versus reduced nicotinamide adenine dinucleotide in melanoma xenografts <1 cm showed strong metabolism in the center of the tumors (32). However, direct quantitation of CCND1 expression in microdissected tumor segments has not yet been performed.
The IGF1-blocking control, the PNA mismatch control [64Cu]WT4322, the peptide mismatch control [64Cu]WT4273, and free 64CuCl2 all showed low levels of 64Cu in the xenografts, which we ascribe to nonspecific extravasation into the xenograft tissue due to increased capillary permeability. Distribution of the CCND1 probe [64Cu]WT4348 to the liver and kidney might reflect appreciable levels of murine ccnd1 mRNA (GenBank NM_007631), identical to the human CCND1 (GenBank NM_053056) over the 12 nucleotides of radiohybridization probe complementarity.
Although the reported stability constant for Cu(II)-DO3A of 2 × 1024 (22) makes transchelation of 64Cu to abundant Cu-binding proteins unlikely, Cu(II) has been asserted to dissociate from DOTA or 1,4,8,11-tetra(carboxymethylaza)cyclotetradecane (TETA) in vivo and bind to superoxide dismutase and metallothioneins in the liver, and to albumin in blood, enabling high 64Cu uptake in liver, blood, and intestine and long retention (33). Our data, however, revealed that 100-fold molar excesses of cysteine, DTPA, or human serum albumin did not displace 64Cu(II) from the [64Cu]DO3A-PNA-peptides. Furthermore, SDS-PAGE analysis of serum isolated from a mouse after administration of the CCND1 probe [64Cu]WT4348 showed no 64Cu radioactivity at 30 kDa, the mass of Cu/Zn superoxide dismutase, which is stable on SDS-PAGE (27).
In addition, tissue distributions with [64Cu]DO3A-PNA-peptides were much lower than tissue distribution of free 64CuCl2 in all tissues except spleen and tumor. Indeed, the %ID/g of free ionic 64Cu(II) in the blood was significantly higher than that of [64Cu]WT4348, implying that tumor uptake of free ionic 64Cu(II) was nonspecific, dependent only on tumor vascularity. Our observations therefore disagree with the hypothesis of 64Cu(II) transchelation.
Comparison of 64Cu PET with 99mTc Scintigraphic Imaging
The microPET studies indicated that the CCND1 probe [64Cu]WT4348 exhibited faster and higher tumor uptake than the corresponding scintigraphic CCND1 probe [99mTc]WT4185 used in our previous study (20) and enhanced in vivo detection, comparable to our earlier results with VPAC1 receptor-specific vasoactive intestinal peptide ligands (26,34). We propose that faster tumor accumulation resulted from the chemical properties of [64Cu(II)-DO3A]−, compared with [99mTc]AcGly-d(Ala)-Gly-Gly (19), which is also subject to oxidation of reduced, chelated 99mTc to 99mTc(VII) (26).
Clinical Translation
Preclinical safety studies in immunocompetent mice of a MYCC PNA-Tat peptide administered intravenously at 0.05 mg/kg found no IgG or IgM antibodies, unless the PNA-peptide was conjugated to keyhole limpet hemocyanin (35). Furthermore, the PNA-peptide was not detectably toxic (36), mutagenic, or clastogenic (37). For clinical imaging of cancer gene mRNA overexpression in breast lesions, a radioprobe should exhibit low lung accumulation, as we have seen for [64Cu]WT4348.
Although liver and kidney accumulation will not interfere with breast imaging, it raises the question of radiation dosimetry. [64Cu]WT4348 has chemical properties and tissue distribution similar to those of [64Cu]TETA-octreotide, which has been administered safely to humans at doses up to 111 MBq (3 mCi), where the bladder wall was the dose-limiting organ (38). In a preclinical study, [64Cu]DO3A-biotin distribution to human colorectal carcinoma xenografts pretargeted with a streptavidin conjugate of the monoclonal antibody NR-LU-10 was compared with that of [64Cu]DO3A-NR-LU-10 binding directly to xenograft cells and other tissues. The small molecule [64Cu]DO3A-biotin, comparable to [64Cu]WT4348, displayed greater tumor accumulation and more rapid blood clearance than the much larger [64Cu]monoclonal antibody (39). Those results imply that a small molecule [64Cu]probe will exhibit greater safety than a [64Cu]monoclonal antibody. We anticipate administration of ≤111 MBq (≤3 mCi) of [64Cu]WT4348 to clinical trial participants, thereby minimizing potential radiotoxicity to normal organs. For clinical trials with [64Cu]WT4348, it will be most practical to administer the radiohybridization probes early in the morning and then image the patients before the end of the day, 6–8 h later, to avoid an overnight stay.
CONCLUSION
We propose that the CCND1 radiohybridization probe could provide noninvasive stratification of radiographically suspect breast masses based on molecular imaging of their genetic profiles. Rapid, noninvasive identification of oncogene activity in mammographic masses or occult lesions might accelerate staging and guide appropriate therapy. Radiohybridization imaging might also identify and stratify malignancies of the prostate, colon, and lung, which have well-established risk factors and biomarkers. Genetic profiling of malignancies by PET might direct the most effective intervention at the onset of disease and enable accurate monitoring of the efficacy of treatment.
Acknowledgments
We thank Dr. Richard Wassell for assistance in measuring matrix-assisted laser desorption/ionization mass spectrometry mass spectra of DO3A-PNA-peptides. The Jefferson Molecular and Biomedical Imaging Core Facility is supported by the Department of Radiology. This work was supported by grants DOE/BER ER63055 and NIH/NCI CA109231.
Footnotes
- COPYRIGHT © 2007 by the Society of Nuclear Medicine, Inc.
References
- 1.↵
Hinds PW, Dowdy SF, Eaton EN, Arnold A, Weinberg RA. Function of a human cyclin gene as an oncogene. Proc Natl Acad Sci USA. 1994;91:709–713. - 2.↵
Weinstat-Saslow D, Merino MJ, Manrow RE, et al. Overexpression of cyclin D mRNA distinguishes invasive and in situ breast carcinomas from non-malignant lesions. Nat Med. 1995;1:1257–1260. - 3.↵
Millikan R, Hulka B, Thor A, et al. p53 mutations in benign breast tissue. J Clin Oncol. 1995;13:2293–2300. - 4.↵
Shoker BS, Jarvis C, Davies MP, Iqbal M, Sibson DR, Sloane JP. Immunodetectable cyclin D(1) is associated with oestrogen receptor but not Ki67 in normal, cancerous and precancerous breast lesions. Br J Cancer. 2001;84:1064–1069. - 5.↵
Surmacz E. Function of the IGF-I receptor in breast cancer. J Mammary Gland Biol Neoplasia. 2000;5:95–105. - 6.↵
Armengol G, Knuutila S, Lluis F, Capella G, Miro R, Caballin MR. DNA copy number changes and evaluation of MYC, IGF1R, and FES amplification in xenografts of pancreatic adenocarcinoma. Cancer Genet Cytogenet. 2000;116:133–141. - 7.↵
Wang Y, Sun Y. Insulin-like growth factor receptor-1 as an anti-cancer target: blocking transformation and inducing apoptosis. Curr Cancer Drug Targets. 2002;2:191–207. - 8.↵
Bhaumik S, Walls Z, Puttaraju M, Mitchell LG, Gambhir SS. Molecular imaging of gene expression in living subjects by spliceosome-mediated RNA trans-splicing. Proc Natl Acad Sci USA. 2004;101:8693–8698. - 9.↵
Wickstrom E. Clinical Trials of Genetic Therapy with Antisense DNA and DNA Vectors. New York, NY: Marcel Dekker; 1998. - 10.↵
- 11.↵
Good L, Nielsen PE. Progress in developing PNA as a gene-targeted drug. Antisense Nucleic Acid Drug Dev. 1997;7:431–437. - 12.↵
Tian X, Wickstrom E. Continuous solid-phase synthesis and disulfide cyclization of peptide-PNA-peptide chimeras. Org Lett. 2002;4:4013–4016. - 13.↵
Gray GD, Basu S, Wickstrom E. Transformed and immortalized cellular uptake of oligodeoxynucleoside phosphorothioates, 3′-alkylamino oligodeoxynucleotides, 2′-O-methyl oligoribonucleotides, oligodeoxynucleoside methylphosphonates, and peptide nucleic acids. Biochem Pharmacol. 1997;53:1465–1476. - 14.↵
Soomets U, Hallbrink M, Langel U. Antisense properties of peptide nucleic acids. Front Biosci. 1999;4:D782–D786. - 15.↵
Basu S, Wickstrom E. Synthesis and characterization of a peptide nucleic acid conjugated to a D-peptide analog of insulin-like growth factor 1 for increased cellular uptake. Bioconjug Chem. 1997;8:481–488. - 16.↵
Pietrzkowski Z, Wernicke D, Porcu P, Jameson BA, Baserga R. Inhibition of cellular proliferation by peptide analogues of insulin-like growth factor 1. Cancer Res. 1992;52:6447–6451. - 17.↵
Rao PS, Tian X, Qin W, et al. 99mTc-Peptide-peptide nucleic acid probes for imaging oncogene mRNAs in tumours. Nucl Med Commun. 2003;24:857–863. - 18.↵
Guvakova MA, Surmacz E. Overexpressed IGF-I receptors reduce estrogen growth requirements, enhance survival, and promote E-cadherin-mediated cell-cell adhesion in human breast cancer cells. Exp Cell Res. 1997;231:149–162. - 19.↵
Soule HD, McGrath CM. Estrogen responsive proliferation of clonal human breast carcinoma cells in athymic mice. Cancer Lett. 1980;10:177–189. - 20.↵
Tian X, Aruva MR, Qin W, et al. External imaging of CCND1 cancer gene activity in experimental human breast cancer xenografts with 99mTc-peptide-peptide nucleic acid-peptide chimeras. J Nucl Med. 2004;45:2070–2082. - 21.↵
Tian X, Aruva MR, Qin W, et al. Noninvasive molecular imaging of MYC mRNA expression in human breast cancer xenografts with a [99mTc]peptide-peptide nucleic acid-peptide chimera. Bioconjug Chem. 2005;16:70–79. - 22.↵
Smith SV. Molecular imaging with copper-64. J Inorg Biochem. 2004;98:1874–1901. - 23.↵
Riesen A, Zehnder M, Kaden TA. Structure of the barium salt of a Cu2+ complex with a tetraaza macrocyclic tetraacetate. Acta Crystallogr. 1988;C44:1740–1742. - 24.↵
Huisman MC, Reder S, Weber AW, Ziegler SI, Schwaiger M. Performance evaluation of the Philips MOSAIC small animal PET scanner. Eur J Nucl Med Mol Imaging. 2007;34:532–540. - 25.↵
Daube-Witherspoon ME, Mate JS, Karp JS, Lewitt RM. The application of the row action maximum likelihood algorithm with spherical basis functions to clinical PET imaging. IEEE Trans Nucl Sci. 2001;48:24–30. - 26.↵
Thakur ML, Aruva MR, Gariepy J, et al. PET imaging of oncogene overexpression using 64Cu-vasoactive intestinal peptide (VIP) analog: comparison with 99mTc-VIP analog. J Nucl Med. 2004;45:1381–1389. - 27.↵
Okado-Matsumoto A, Fridovich I. Subcellular distribution of superoxide dismutases (SOD) in rat liver: Cu,Zn-SOD in mitochondria. J Biol Chem. 2001;276:38388–38393. - 28.↵
Tian X, Aruva MR, Rao PS, et al. Imaging oncogene expression. In: Cho-Chung YS, Gewirtz AM, Stein CA, eds. Therapeutic Oligonucleotides: Antisense, RNAi, Triple Helix, DNA Decoys, and DNA Chips. Vol 1002. New York, NY: New York Academy of Sciences; 2003:1083–1099. - 29.↵
Sun X, Fang H, Li X, Rossin R, Welch MJ, Taylor JS. MicroPET imaging of MCF-7 tumors in mice via unr mRNA-targeted peptide nucleic acids. Bioconjug Chem. 2005;16:294–305. - 30.↵
Tufto I, Lyng H, Rofstad EK. Vascular density in human melanoma xenografts: relationship to angiogenesis, perfusion and necrosis. Cancer Lett. 1998;123:159–165. - 31.↵
Medarova Z, Pham W, Farrar C, Petkova V, Moore A. In vivo imaging of siRNA delivery and silencing in tumors. Nat Med. 2007;13:372–377. - 32.↵
Li LZJ, Zhong T, Zhou R, et al. Predicting melanoma metastatic potential by optical and magnetic resonance imaging. In: McGuire DJ, Bruley DF, Harrison DK, eds. Oxygen Transport to Tissue XXVIII. Vol. 599. New York, NY: Springer; 2007:67–78. - 33.↵
Boswell CA, Sun X, Niu W, et al. Comparative in vivo stability of copper-64-labeled cross-bridged and conventional tetraazamacrocyclic complexes. J Med Chem. 2004;47:1465–1474. - 34.↵
Pallela VR, Thakur ML, Chakder S, Rattan S. 99mTc-Labeled vasoactive intestinal peptide receptor agonist: functional studies. J Nucl Med. 1999;40:352–360. - 35.↵
Cutrona G, Boffa LC, Mariani MR, et al. The peptide nucleic acid targeted to a regulatory sequence of the translocated c-myc oncogene in Burkitt's lymphoma lacks immunogenicity: follow-up characterization of PNAEmu-NLS. Oligonucleotides. 2007;17:146–150. - 36.↵
Boffa LC, Cutrona G, Cilli M, et al. Therapeutically promising PNA complementary to a regulatory sequence for c-myc: pharmacokinetics in an animal model of human Burkitt's lymphoma. Oligonucleotides. 2005;15:85–93. - 37.↵
Boffa LC, Cutrona G, Cilli M, et al. Inhibition of Burkitt's lymphoma cells growth in SCID mice by a PNA specific for a regulatory sequence of the translocated c-myc. Cancer Gene Ther. 2007;14:220–226. - 38.↵
Anderson CJ, Dehdashti F, Cutler PD, et al. 64Cu-TETA-octreotide as a PET imaging agent for patients with neuroendocrine tumors. J Nucl Med. 2001;42:213–221. - 39.↵
Lewis MR, Wang M, Axworthy DB, et al. In vivo evaluation of pretargeted 64Cu for tumor imaging and therapy. J Nucl Med. 2003;44:1284–1292.
- Received for publication April 4, 2007.
- Accepted for publication July 30, 2007.