Frontiers | Informatics and Data Mining Tools and Strategies for the Human Connectome Project (original) (raw)
Introduction
The past decade has seen great progress in the refinement of non-invasive neuroimaging methods for assessing long-distance connections in the human brain. This has given rise to the tantalizing prospect of systematically characterizing human brain connectivity, i.e., mapping the connectome (Sporns et al., 2005). The eventual elucidation of this amazingly complex wiring diagram should reveal much about what makes us uniquely human and what makes each person different from all others.
The NIH recently funded two consortia under the Human Connectome Project (HCP)1. One is led by Washington University and University of Minnesota and involves seven other institutions (the “WU-Minn HCP consortium”)2. The other, led by Massachusetts General Hospital and UCLA (the MGH/UCLA HCP consortium), focuses on building and refining a next-generation 3T MR scanner for improved sensitivity and spatial resolution. Here, we discuss informatics aspects of the WU-Minn HCP consortium’s plan to map human brain circuitry in 1,200 healthy young adults using cutting-edge non-invasive neuroimaging methods. Key imaging modalities will include diffusion imaging, resting-state fMRI, task-evoked fMRI, and magnetoencephalography combined with electroencephalography (MEG/EEG). A battery of behavioral and cognitive tests will also be included along with the collection of genetic material. This endeavor will yield valuable information about brain connectivity, its relationship to behavior, and the contributions of genetic and environmental factors to individual differences in brain circuitry. The data generated by the WU-Minn HCP consortium will be openly shared with the scientific community.
The HCP has a broad informatics vision that includes support for the acquisition, analysis, visualization, mining, and sharing of connectome-related data. As it implements this agenda, the consortium seeks to engage the neuroinformatics community through open source software, open programming interfaces, open-access data-sharing, and standards-based development. The HCP informatics approach includes three basic domains.
• Data support components include tools and services that manage data (e.g., data uploads from scanners and other data collection devices); execution and monitoring of quality assurance, image processing, and analysis pipelines and routines; secure long-term storage of acquired and processed data; search services to identify and select subsets of the data; and download mechanisms to distribute data to users around the globe.
• Visualization components include a spectrum of tools to view anatomic and functional brain data in volumetric and surface representations and also using network and graph-theoretic representations of the connectome.
• Discovery components are an especially important category of the HCP’s informatics requirements, including user interfaces (UI) for formulating database queries, linking between related knowledge/database systems, and exploring the relationship of an individual’s connectome to population norms.
The HCP is expected to generate approximately 1 PB of data, which will be made accessible via a tiered data-sharing strategy. Besides the sheer amount of data, there will be major challenges associated with handling the diversity of data types derived from the various modalities of data acquisition, the complex analysis streams associated with each modality, and the need to cope with individual variability in brain shape as well as brain connectivity, which is especially dramatic for cerebral cortex.
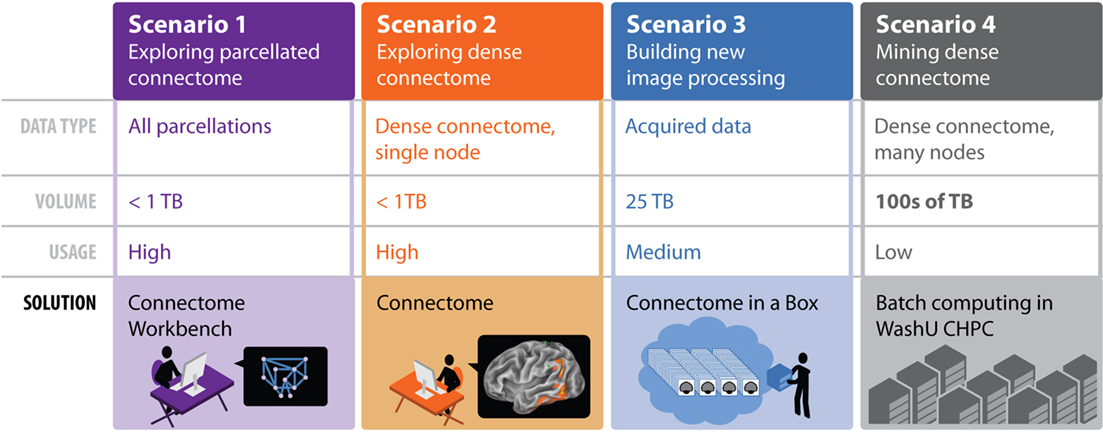
To support these needs, the HCP is developing a comprehensive informatics platform centered on two interoperable components: ConnectomeDB, a data management system, and Connectome Workbench (CWB), a software suite that provides visualization and discovery capabilities.
ConnectomeDB is based on the XNAT imaging informatics platform, a widely used open source system for managing and sharing imaging and related data (Marcus et al., 2007)3. XNAT includes an open web services application programming interface (API) that enables external client applications to query and exchange data with XNAT hosts. This API will be leveraged within the HCP informatics platform and will also help externally developed applications connect to the HCP. CWB is based on Caret software, a visualization and analysis platform that handles structural and functional data represented on surfaces and volumes and on individuals and atlases (Van Essen et al., 2001). The HCP also benefits from a variety of processing and analysis software tools, including FreeSurfer, FSL, and FieldTrip.
Here, we provide a brief overview of the HCP, then describe the HCP informatics platform in some detail. We also provide a sampling of the types of scientific exploration and discovery that it will enable.
Overview of the Human Connectome Project
Inferring Long-Distance Connectivity from in vivo Imaging
The two primary modalities for acquiring information about human brain connectivity in vivo are diffusion imaging (dMRI), which provides information about structural connectivity, and resting-state functional MRI (R-fRMI), which provides information about functional connectivity. The two approaches are complementary, and each is very promising. However, each has significant limitations that warrant brief comment.
Diffusion imaging relies on anisotropies in water diffusion to determine the orientation of fiber bundles within white matter. Using High Angular Resolution Diffusion Imaging (HARDI), multiple fiber orientations can be identified within individual voxels. This enables tracking of connections even in regions where multiple fiber bundles cross one another. Probabilistic tractography integrates information throughout the white matter and can reveal detailed information about long-distance connectivity patterns between gray-matter regions (Johansen-Berg and Behrens, 2009; Johansen-Berg and Rushworth, 2009). However, uncertainties arising at different levels of analysis can lead to both false positives and false negatives in tracking connections. Hence, it is important to continue refining the methods for dMRI data acquisition and analysis.
R-fMRI is based on spatial correlations of the slow fluctuations in the BOLD fMRI signal that occur at rest or even under anesthesia (Fox and Raichle, 2007). Studies in the macaque monkey demonstrate that R-fMRI correlations tend to be strong for regions known to be anatomically interconnected, but that correlations can also occur between regions that are linked only indirectly (Vincent et al., 2007). Thus, while functional connectivity maps are not a pure indicator of anatomical connectivity, they represent an invaluable measure that is highly complementary to dMRI and tractography, especially when acquired in the same subjects.
The HCP will carry out a “macro-connectome” analysis of long-distance connections at a spatial resolution of 1–2 mm. At this scale, each gray-matter voxel contains hundreds of thousands of neurons and hundreds of millions of synapses. Complementary efforts to chart the “micro-connectome” at the level of cells, dendrites, axons, and synapses aspire to reconstruct domains up to a cubic millimeter (Briggman and Denk, 2006; Lichtman et al., 2008), so that the macro-connectome and micro-connectome domains will barely overlap in their spatial scales.
A Two-Phase HCP Effort
Phase I of the 5-year WU-Minn HCP consortium grant is focused on additional refinements and optimization of data acquisition and analysis stages and on implementing a robust informatics platform. Phase II, from mid-2012 through mid-2015, will involve data acquisition from the main cohort of 1,200 subjects as well as continued refinement of the informatics platform and some analysis methods. This section summarizes key HCP methods relevant to the informatics effort and describes some of the progress already made toward Phase I objectives. A more detailed description of our plans will be published elsewhere.
Subject Cohort
We plan to study 1,200 subjects (300 healthy twin pairs and available siblings) between the ages of 22 and 35. This design, coupled with collection of subjects’ DNA, will yield invaluable information about (i) the degree of heritability associated with specific components of the human brain connectome; and (ii) associations of specific genetic variants with these components in healthy adults. It will also enable genome-wide testing for additional associations (e.g., Visscher and Montgomery, 2009).
Imaging
All 1,200 subjects will be scanned at Washington University on a dedicated 3 Tesla (3T) Siemens Skyra scanner. The scanner will be customized to provide a maximum gradient strength of ∼100 mT/m, more than twice the standard 40 mT/m for the Skyra. A subset of 200 subjects will also be scanned at the University of Minnesota using a new 7T scanner, which is expected to provide improved signal-to-noise ratio and better spatial resolution, but is less well established for routine, high-throughput studies. Some subjects may also be scanned on a 10.5 T scanner currently under development at the University of Minnesota. Having higher-field scans of individuals also scanned at 3T will let us use the higher-resolution data to constrain and better interpret the 3T data.
Each subject will have multiple MR scans, including HARDI, R-fMRI (Resting-state fMRI), T-fMRI (task-evoked fMRI), and standard T1-weighted and T2-weighted anatomical scans. Advances in pulse sequences are planned in order to obtain the highest resolution and quality of data possible in a reasonable period of time. Already, new pulse sequences have been developed that accelerate image acquisition time (TR) by sevenfold while maintaining or even improving the signal-to-noise ratio (Feinberg et al., 2010). The faster temporal resolution for both R-fMRI and T-fMRI made possible by these advances will increase the amount of data acquired for each subject and increase the HCP data storage requirements, a point that exemplifies the many interdependencies among various HCP project components.
Task-fMRI scans will include a range of tasks aimed at providing broad coverage of the brain and identifying as many functionally distinct parcels as possible. The results will aid in validating and interpreting the results of the connectivity analyses obtained using resting-state fMRI and diffusion imaging. These “functional localizer” tasks will include measures of primary sensory processes (e.g., vision, motor function) and a wide range of cognitive and affective processes, including stimulus category representations, working memory, episodic memory, language processing, emotion processing, decision-making, reward processing and social cognition. The specific tasks to be included are currently being piloted; final task selection will be based on multiple criteria, including sensitivity, reliability and brain coverage.
A subset of 100 subjects will also be studied with combined MEG/EEG, which provides vastly better temporal resolution (milliseconds instead of seconds) but lower spatial resolution than MR (between 1 and 4 cm). Mapping MEG/EEG data to cortical sources will enable electrical activity patterns among neural populations to be characterized as functions of both time and frequency. As with the fMRI, MEG/EEG will include both resting-state and task-evoked acquisitions. The behavioral tasks will be a matched subset of the tasks used in fMRI. The MEG/EEG scans, to will be acquired at St. Louis University using a Magnes 3600 MEG (4DNeuroimaging, San Diego, CA, USA) with 248 magnetometers, 23 MEG reference channels (5 gradiometer, and 18 magnetometer) and 64 EEG voltage channels. This data will be analyzed in both sensor space and using state-of-the-art source localization methods (Wipf and Nagarajan, 2009; Ou et al., 2010) and using subject specific head models derived from anatomic MRI. Analyses of band-limited power (BLP) will provide measures that reflect the frequency-dependent dynamics of resting and task-evoked brain activity (de Pasquale et al., 2010; Scheeringa et al., 2011).
Behavioral, Genetic, and Other Non-Imaging Measures
Measuring behavior in conjunction with mapping of structural and functional networks in HCP subjects will enable the analysis of the functional correlates of variations in “typical” brain connectivity and function. It will also provide a starting point for future studies that examine how abnormalities in structural and functional connectivity play a role in neurological and psychiatric disorders.
The HCP will use a battery of reliable and well-validated measures that assess a wide range of human functions, including cognition, emotion, motor and sensory processes, and personality. The core of this battery will be from the NIH Toolbox for Assessment of Neurological and Behavioral function4. This will enable federation of HCP data with other large-scale efforts to acquire neuroimaging and behavioral data and will facilitate comparison of brain-behavior relationships across studies (Gershon et al., 2010). Additional tests that are currently being piloted will be drawn from other sources.
Genetic Analyses
Blood samples collected from each subject during their visit will be sent to the Rutgers University Cell and DNA Repository (RUCDR), where cell lines will be created and DNA will be extracted. Genetic analysis will be conducted in early 2015, after all Phase II subjects have completed in-person testing. Performing the genotyping in the later stages of the project will allow the HCP to take advantage of future developments in this rapidly advancing field, including the availability of new sequencing technologies and decreased costs of whole-genome sequencing. Genetic data and de-identified demographic and phenotype data will be entered into the dbGAP database in accordance with NIH data-sharing policies. Summary data look-up by genotype will be possible via ConnectomeDB.
Study Workflow
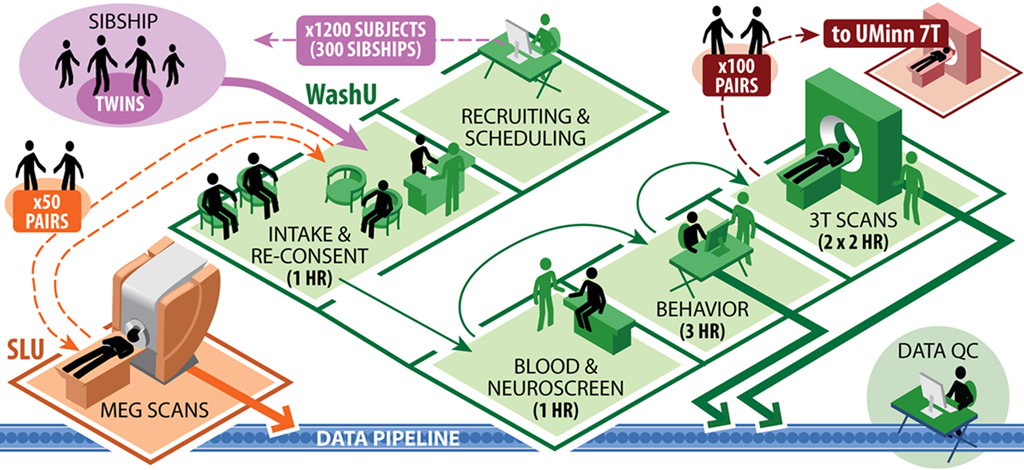
The collection of this broad range of data types from multiple family groups will necessitate careful coordination of the various tests during in-person visits. Figure 1 illustrates the data collection workflow planned for the high-throughput phase of the HCP. All 1,200 subjects in the main cohort will be scanned at Washington University on the dedicated 3T scanner. A subset of 200 subjects (100 same-sex twin pairs, 50% monozygotic) will also be scanned at University of Minnesota using 7T MRI (HARDI, R-fMRI, and T-fMRI) and possibly also 10.5 T. Another subset of 100 (50 same-sex twin pairs, all monozygotic) will be scanned at St. Louis University (SLU) using MEG/EEG. Many data management and quality control (QC) steps will be taken to maximize the quality and reliability of these datasets (see Data Workflow and Quality Control sections).
Figure 1. HCP subject workflow.
The HCP Informatics Approach
Our HCP informatics approach includes components related to data support and visualization. The Section “Data Support” discusses key data types and representations plus aspects of data processing pipelines that have major informatics implications. This leads to a discussion of ConnectomeDB and the computational resources and infrastructure needed to support it, as well as our data-sharing plans. The Section “Visualization” describes CWB and its interoperability with ConnectomeDB. These sections also include examples of potential exploratory uses of HCP data.
Data Support
Data types
Volumes, surfaces, and representations. MR images are acquired in a 3-D space of regularly spaced voxels, but the geometric representations useful for subsequent processing depend upon brain structure. Subcortical structures are best processed in standard volumetric (voxel) coordinates. The complex convolutions of the cortical sheet make it advantageous for many purposes to model the cortex using explicit surface representations – a set of vertices topologically linked into a 2D mesh for each hemisphere. However, for other purposes it remains useful to analyze and visualize cortical structures in volume space. Hence, the HCP will support both volumetric and surface representations for analysis and visualization.
For some connectivity data types, it is useful to represent subcortical volumetric coordinates and cortical surface vertices in a single file. This motivates introduction of a geometry-independent terminology. Specifically, a “_brainordinate_” (brain coordinate) is a spatial location within the brain that can be either a voxel (i, j, k integer values) or a surface vertex (x, y, z real-valued coordinates and a “node number”); a “_grayordinate_” is a voxel or vertex within gray matter (cortical or subcortical); a “_whiteordinate_” is a voxel within white matter or a vertex on the white matter surface. These terms (brainordinate, grayordinate, and whiteordinate) are especially useful in relation to the CIFTI data files described in the next paragraph.
When feasible, the HCP will use standard NIFTI-1 (volumetric) and GIFTI (surfaces) formats. Primary diffusion imaging data will be stored using the format MiND recently developed by Patel et al. (2010). By conforming to these existing formats, datasets generated using one software platform can be read by other platforms without the need to invoke file conversion utilities. Several types of connectivity-related data will exceed the size limits supported by NIFTI-1 and GIFTI and will instead use the recently adopted NIFTI-2 format5. NIFTI-2 is similar to NIFTI-1, but has dimension indices increased from 16-bit to 64-bit integers, which will be useful for multiple purposes and platforms. For the HCP, connectivity or time-series values will be stored in the binary portion of the NIFTI-2 format. Datasets whose brainordinates include both voxels and surface vertices pose special metadata requirements that are being addressed for the HCP and for other software platforms by a “CIFTI” working group (with “C” indicating connectivity). A description of CIFTI data types including example file formats has been reviewed by domain experts and is available for public comment6. CIFTI file formats will support metadata that map matrix rows and columns to brainordinates, parcels (see below), and/or time points, in conformance with NIFTI conventions for header extensions.
Individuals, atlases, and registration. The anatomical substrates on which HCP data are analyzed and visualized will include individual subjects as well as atlases. In general, quantitative comparisons across multiple subjects require registering data from individuals to an atlas. Maximizing the quality of inter-subject registration (alignment) is a high priority but also a major challenge. This is especially the case for cerebral cortex, owing to the complexity and variability of its convolutions. Several registration methods and atlases are under consideration for the HCP, including population-average volumes and population-average cortical surfaces based on registration of surface features. Major improvements in inter-subject alignment may be attainable by invoking constraints related to function, architecture, and connectivity, especially for cerebral cortex (e.g., Petrovic et al., 2007; Sabuncu et al., 2010). This is important for the HCP informatics effort, insofar as improved atlas representations that emerge in Phase II may warrant support by the HCP.
Parcellations. The brain can be subdivided into many subcortical nuclei and cortical areas (“_parcels_”), each sharing common characteristics based on architectonics, connectivity, topographic organization, and/or function. Expression of connectivity data as a matrix of connection weights between parcels will enable data to be stored very compactly and transmitted rapidly. Also, graph-theoretic network analyses (see below) will be more tractable and biologically meaningful on parcellated data. However, this will place a premium on the fidelity of the parcellation schemes. Data from the HCP should greatly improve the accuracy with which the brain can be subdivided, but over a time frame that will extend throughout Phase II. Hence, just as for atlases, improved parcellations that emerge in Phase II may warrant support by the HCP.
Networks and modularity. Brain parcels can often be grouped into spatially distributed networks and subnetworks that subserve distinct functions. These can be analyzed using graph-theoretic approaches that model networks as nodes connected by edges (Sporns, 2010). In the context of HCP, graph nodes can be brainordinates or parcels, and edges can be R-fMRI correlations (full correlations or various types of partial correlations), tractography-based estimates of connection probability or strength, or other measures of relationships between the nodes. The HCP will use several categories of network-related measures, including measures of segregation such as clustering and modularity (Newman, 2006); measures of integration, including path length and global efficiency; and measures of influence to identify subsets of nodes and edges central to the network architecture such as hubs or bridges (Rubinov and Sporns, 2010).
Processing pipelines and analysis streams. Generation of the various data types for each of the major imaging modalities will require extensive processing and analysis. Each analysis stream needs to be carried out in a systematic and well-documented way. For each modality, a goal is to settle on customized processing streams that yield the highest-quality and most informative types of data. During Phase I, this will include systematic evaluation of different pipelines and analysis strategies applied to the same sets of preliminary data. Minimally processed versions for each data modality will also remain available, which will enable investigators to explore alternative processing and analysis approaches.
ConnectomeDB
XNAT foundation. ConnectomeDB is being developed as a customized version of the XNAT imaging informatics platform (Marcus et al., 2007). XNAT is a highly extensible, open source system for receiving, archiving, managing, processing, and sharing both imaging and non-imaging study data. XNAT includes five services that are critical for ConnectomeDB operations. The DICOM Service receives and stores data from DICOM devices (scanners or gateways), imports relevant metadata from DICOM tags to the database, anonymizes sensitive information in the DICOM files, and converts the images to NIFTI formatted files. The Pipeline Service for defining and executing automated and semi-automated image processing procedures allows computationally intensive processing and analysis jobs to be offloaded to compute clusters while managing, monitoring and reporting on the execution status of these jobs through its application interface. The Quality Control Service enables both manual and automated review of images and subsequent markup of specific characteristics (e.g., motion artifacts, head positioning, signal to noise ratio) and overall usability of individual scans and full imaging sessions. The Data Service allows study data to be incorporated into the database. The default data model includes a standard experiment hierarchy, including projects, subjects, visits, and experiments. On top of this basic hierarchy, specific data type extensions can be added to represent specific data, including imaging modalities, derived imaging measures, behavioral tests, and genetics information. The Data Service provides mechanisms for incorporating these extensions into the XNAT infrastructure, including the database backend, middleware data access objects, and frontend reports and data entry forms. Finally, the Search Service allows complex queries to be executed on the database.
All of XNAT’s services are accessible via an open web services API that follows the REpresentational State Transfer (REST) approach (Fielding, 2000). By utilizing the richness of the HTTP protocol, REST web services allow requests between client and server to be specified using browser-like URLs. The REST API provides specific URLs to create, access, and modify every resource under XNAT’s management. The URL structures follow the organizational hierarchy of XNAT data, making it intuitive to navigate the API either manually (rarely) or programmatically. HCP will use this API for interactions between ConnectomeDB and CWB, for importing data into and out of processing pipelines, and as a conduit between external software applications and HCP datasets. External libraries and tools that can interact with the XNAT API include pyxnat – a Python library for interfacing with XNAT repositories7; 3D Slicer – an advanced image visualization and analysis environment8; and LONI Pipeline – a GUI-based pipelining environment9.
API extensions. The HCP is developing additional services to support connectome-related queries. A primary initial focus is on a service that enables spatial queries on connectivity measures. This service will calculate and return a connectivity map or a task-evoked activation map based on specified spatial, subject, and calculation parameters. The spatial parameter will allow queries to specify the spatial domain to include in the calculation. Examples include a single brainordinate (see above), a cortical or subcortical parcel, or some other region of interest (collection of brainordinates). This type of search will benefit from registering each subject’s data onto a standard surface mesh and subcortical atlas parcellation. The subject parameter will allow queries to specify the subject or subject groups to include in the calculation examples including an individual subject ID, one or more lists of subject IDs, subject characteristics (e.g., subjects with IQ > 120, subjects with a particular genotype at a particular genetic locus), and contrasts (e.g., subjects with IQ > 110 vs. subjects with IQ < 90). Finally, the calculation parameter will allow queries to specify the specific connectivity or task-evoked activation measure to calculate and return. Basic connectivity measures will include those based on resting-state fMRI (functional connectivity) and diffusion imaging (structural connectivity). Depending on the included subject parameter, the output connectivity measure might be the individual connectivity maps for a specific subject, the average map for a group of subjects, or the average difference map between two groups. When needed, the requested connectivity information (e.g., average difference maps) will be dynamically generated. Task-evoked activation measures will include key contrasts for each task and options to they view activation maps for a particular task in a specific subject, the average map for a group of subjects, or comparing two groups.
Importantly, connectivity results will be accessible either as dense connectivity maps, which will have fine spatial resolution but will be slower to compute and transmit, or as parcellated connectivity maps, which will be faster to process and in some situations may be pre-computed. Additional features that are planned include options to access time courses for R-fMRI data, fiber trajectories for structural connectivity data, and individual subject design files and time courses for T-fMRI data. Other approaches such as regression analysis will also be supported. For example, this may include options to determine the correlation between features of particular pathways or networks and particular behavioral measures (e.g., working memory).
When a spatial query is submitted, ConnectomeDB will parse the parameters, search the database to identify the appropriate subjects, retrieve the necessary files from its file store, and then execute the necessary calculations. By executing these queries on the database server and its associated computing cluster, only the final connectivity or activation map will need to be transferred back to the user. While this approach increases the computing demands on the HCP infrastructure, it will dramatically reduce the amount of data that needs to be transferred over the network. CWB will be a primary consumer of this service, but as with all services in the ConnectomeDB API, it will be accessible to other external clients, including other visualization environments and related databases.
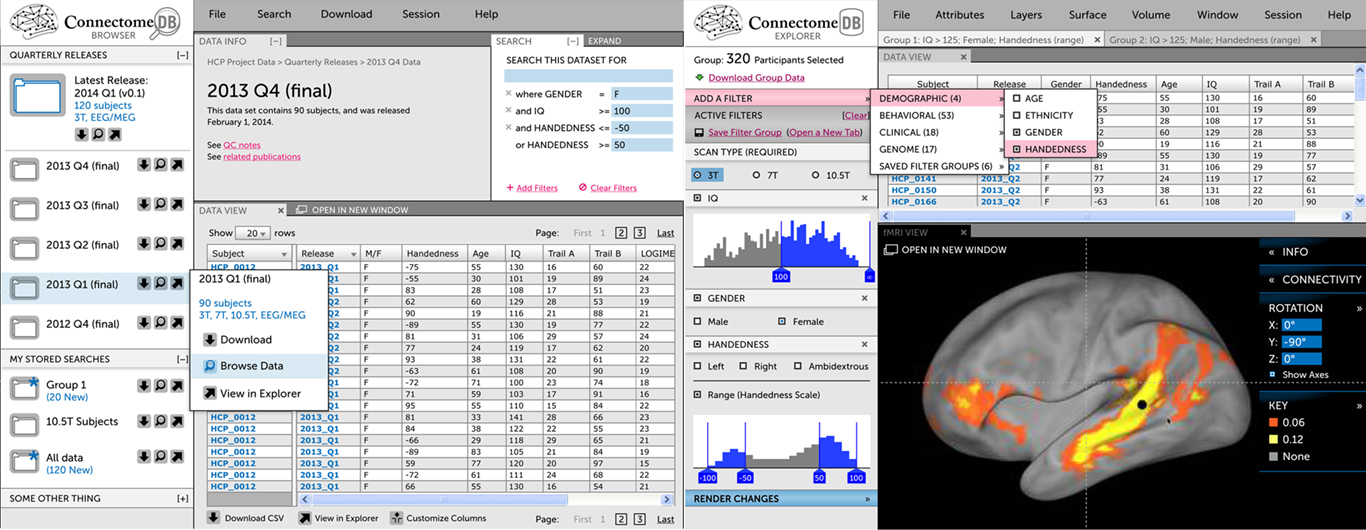
User interface. The ConnectomeDB UI is being custom developed using dynamic web technologies (HTML 5, Javascript, Ajax; Figure 2). Building on advanced web technologies has several advantages, including streamlined access to remote data, high levels of dynamic user interaction, and portability across client systems (browsers, desktop applications, mobile devices). The interface will include two main tracks. The Download track emphasizes rapid identification of data of interest and subsequent download. The most straightforward downloads will be pre-packaged bundles, containing high interest content from each quarterly data release (see Data-Sharing below). Alternatively, browsing and search interfaces will allow users to select individual subjects and subjects groups by one or more demographic, genetic, or behavioral criteria. The Visualization & Discovery track will include an embedded version of CWB, which will allow users to explore connectivity data on a rendered 3D surface (see Visualization below). Using a faceted search interface, users will build subject groups that are dynamically rendered by CWB.
Figure 2. The Connectome UI. (Left) This mockup of the Visualization & Discovery track illustrates key concepts that are being implemented, including a faceted search interface to construct subject groups and an embedded version of Connectome Workbench. Both the search interface and Workbench view are fed by ConnectomeDB’s open API. (Right) This mockup of the Download track illustrates the track’s emphasis on guiding users quickly to standard download packages and navigation to specific data.
High-throughput informatics
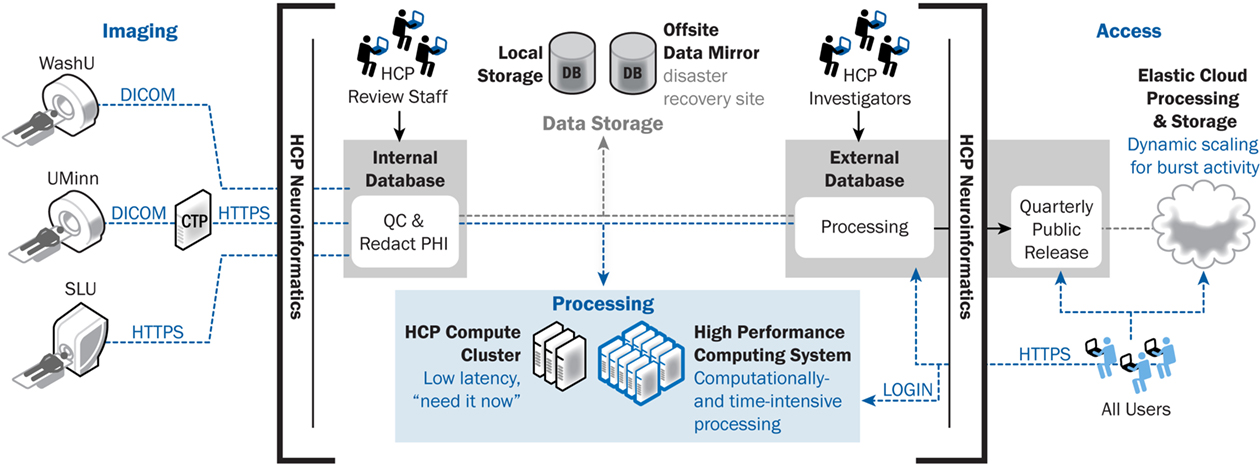
The HCP informatics platform will support high-throughput data collection and open-access data-sharing. Data collection requirements include uploading acquired data from multiple devices and study sites, enforcing rigorous QC procedures, and executing standardized image processing. Data-sharing requirements include supporting world-wide download of very large data sets and high volumes of API service requests. The overall computing and database strategy for supporting these requirements is illustrated in Figure 3 and detailed below.
Figure 3. ConnectomeDB architecture, including data transfer components. ConnectomeDB will utilize the Tomcat servlet container as the application server and use the enterprise grade, open source PostgreSQL database for storage of non-imaging data, imaging session meta-data, and system data. Actual images and other binary content are stored on a file system rather than in the database, improving performance and making the data more easily consumable by external software packages.
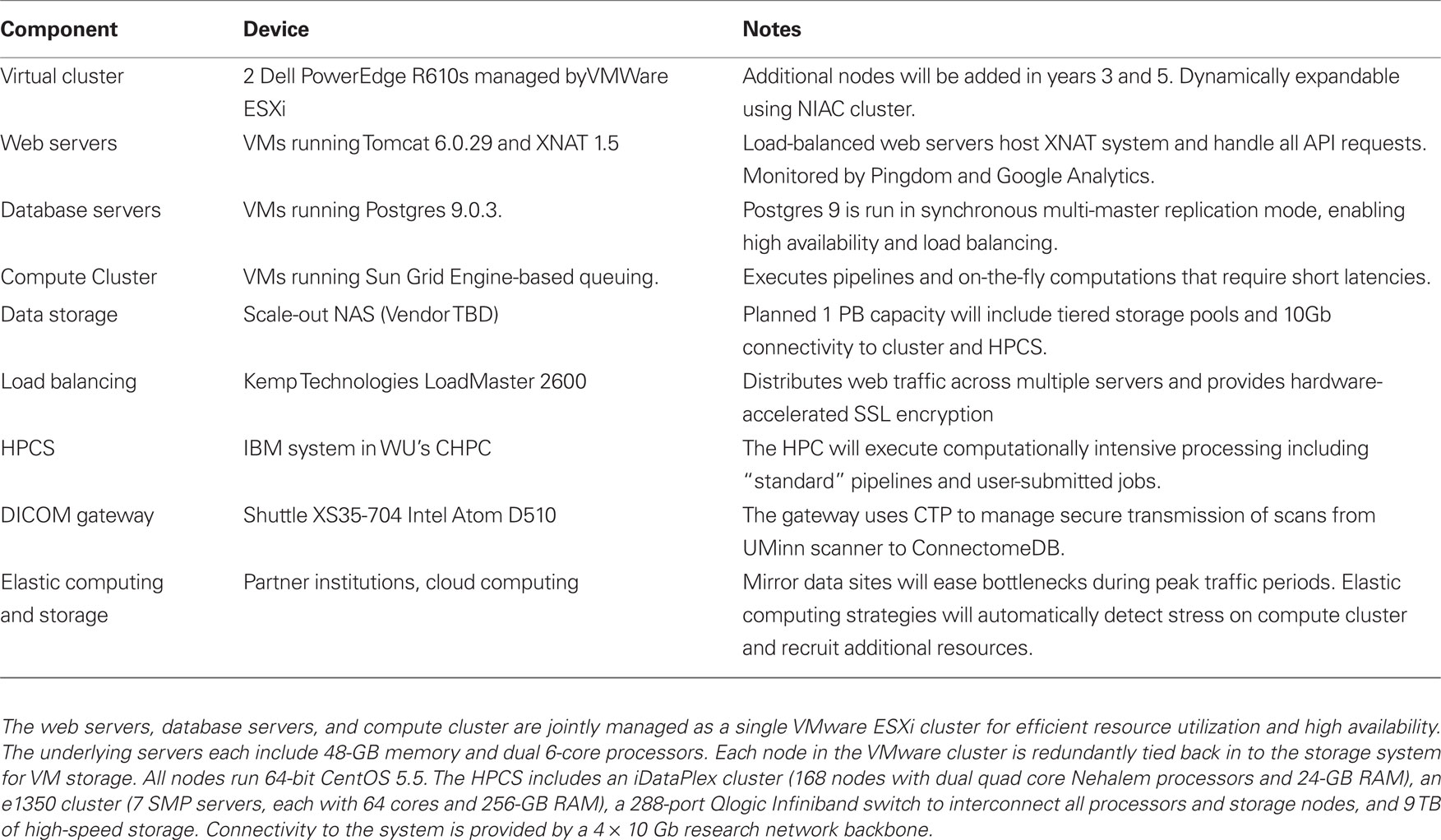
Computing infrastructure. The HCP computing infrastructure (Table 1) includes two complementary systems, an elastically expandable virtual cluster and a high performance computing system (HPCS). The virtual cluster has a pool of general purpose servers managed by VMW are ESXi. Specific virtual machines (VMs) for web servers, database servers, and compute nodes are allocated from the VMW are cluster and can be dynamically provisioned to match changing load conditions. Construction of the VMs is managed by Puppet (Puppet Labs), a systems management platform that enables IT staff to manage and deploy standard system configurations. The initial Phase 1 cluster includes 4 6-core physical CPUs that will be expanded in project years 3 and 5. We will partner with the WU Neuroimaging Informatics and Analysis Center (NIAC), which runs a similar virtual cluster, to dynamically expand the HCP’s capacity during peak load. During extremely high load, we may also utilize commercial cloud computing services to elastically expand the cluster’s computing capacity.
Table 1. The HCP computing infrastructure.
To support the project’s most demanding processing streams, we have partnered with the WU Center for High Performance Computing (CHPC), which operates an IBM HPCS that commenced operating in 2010. Pipelines developed for the HCP greatly benefit from the ability to run in parallel across subjects and take advantage of the vast amount of memory available in the HPCS nodes. Already, several neuroimaging packages including FreeSurfer, FSL, and Caret have been installed on the platform and are in active use by the HCP. The system utilizes a MOAB/TORQUE scheduling system that manages job priority. While the CHPC’s HPCS is a shared resource openly available to the University’s research community, the HCP will have assured priority on the system to ensure that the project has sufficient resources to achieve its goals.
The two HCP computing systems are complementary in that the virtual cluster provides rapid response times and can be dynamically expanded to match load. The HPCS, on the other hand, has large computing power but is a shared resource that queues jobs. The virtual cluster is therefore best for on-the-fly computing, such as is required to support web services, while the HPCS is best for computationally intensive pipelines that are less time sensitive.
The total volume of data produced by the HCP will likely be multiple petabytes (1 petabyte = 1,000,000 gigabytes). We are currently evaluating data storage solutions that handle data at this scale to determine the best price/performance ratio for the HCP. Based on preliminary analyses, we are expecting to deploy 1 PB of storage, which will require significant compromises in deciding which of the many data types generated will be preserved. Datasets to be stored permanently will include primary data plus the outputs of key pre-processing and analysis stages. These will be selected on the basis of their expected utility to the community and on the time that would be needed to recompute or regenerate intermediate processing results.
A driving consideration in selecting a storage solution is close integration with the HPCS. Four 10-Gb network connections between the two systems will enable high-speed data transmission, which will put serious strain on the storage device. Given these connections and the HPCS’s architecture, at peak usage, the storage system will need to be able to sustain up to 200,000 input/output operations per second, a benchmark achievable by a number of available scale-out NAS (Network Attached Storage) systems. To meet this benchmark, we expect to design a system that includes tiered storage pools with dynamic migration between tiers.
In addition to this core storage system, we are also planning for backup, disaster recovery, and mirror sites. Given the scale of the data, it will be impossible to backup all of the data, so we will prioritize data that could not be regenerated, including the raw acquired data and processed data that requires significant computing time. We will utilize both near-line backups for highest priority data and offsite storage for catastrophic disaster recovery. As described below, our data-sharing plan includes quarterly data releases throughout Phase 2. To reduce bottlenecks during peak periods after these releases, we aim to mirror the current release on academic partner sites and commercial cloud systems. We are also exploring distribution through the BitTorrent model (Langille and Eisen, 2011).
Data workflow. All data acquired within the HCP will be uploaded or entered directly into ConnectomeDB. ConnectomeDB itself includes two separate database systems. Initially, data are entered into an internal-facing system that is accessible only to a small group of HCP operations staff who are responsible for reviewing data quality and project workflow. Once data pass quality review, they will be de-identified, including removal of sensitive fields from the DICOM headers and obscuring facial features in the high-resolution anatomic scans, transferred to a public-facing database, and shared with the public according to the data-sharing plan described below. All processing and analysis pipelines will be executed on the public-facing system so that these operations are performed on de-identified data only.
MRI data acquired at Washington University will be uploaded directly from the scanner to ConnectomeDB over the DICOM protocol on a secure private network. MRI data acquired at the University of Minnesota will be sent from the scanners to an on-site DICOM gateway configured with RSNA’s Clinical Trial Processor (CTP) software. The CTP appliance will receive the data over the DICOM protocol, which is non-encrypted, and relay it to ConnectomeDB over the secure HTTPS protocol. Once the data have been uploaded, several actions will be triggered. First, XNAT’s DICOM service will import metadata from the DICOM header fields into the database and places the files into its file repository. Next, a notification will be sent to HCP imaging staff to complete manual inspection of the data. Finally, a series of pipelines will be executed to generate sequence-specific automated QC metrics with flags to the HCP imaging staff regarding problematic data, and to validate metadata fields for protocol compliance. We aim to complete both manual and automated QA within 1 h of acquisition, which will enable re-scanning of individuals while they are still on-site.
MEG/EEG data will be uploaded to ConnectomeDB via a dedicated web form in native 4D format that will insure de-identification and secure transport via https. QC procedures will ensure proper linkage to other information via study specific subject IDs. EEG data will be converted to European Data format (EDF)10 while MEG data will remain in source format.
Demographic and behavioral data will be entered into ConnectomeDB, either through import mechanisms or direct data entry. Most of the behavioral data will be acquired on the NIH Toolbox testing system, which includes its own database. Scripts are being developed to extract the test results from the Toolbox database and upload them into ConnectomeDB via XML documents. Additional connectome-specific forms will be developed for direct web-based entry into ConnectomeDB, via desktop or tablet computers.
Quality control. Initial QC of imaging data will be performed by the technician during acquisition of the data by reviewing the images at the scanner console. Obviously flawed data will be immediately reacquired within the scan session. Once imaging studies have been uploaded to the internal ConnectomeDB, several QC and pre-processing procedures will be triggered and are expected to be completed within an hour, as discussed above. First, the scans will be manually inspected in more detail by trained technicians. The manual review process will use a similar procedure as that used by the Alzheimer’s Disease Neuroimaging Initiative, which includes evaluation of head positioning, susceptibility artifacts, motion, and other acquisition anomalies along a 4-point scale (Jack et al., 2008). Specific extensions will be implemented for BOLD and diffusion imaging. Second, automated programs will be run to assess image quality. Specific quality metrics are currently being developed for each of the HCP imaging modalities and behavioral paradigms. The resulting metrics will be compared with the distribution of values from previous acquisitions to determine whether each is within an expected range. During the initial months of data acquisition, the number of HCP scans contributing to these norm values will be limited, so we will seed the database with values extracted from data obtained in similar studies and during the pilot phase. As the study database expands, more sophisticated approaches will become available, including metrics specific for individual fMRI tasks (which may vary in the amount of head motion). Specific QC criteria for each metric will be developed during Phase I.
Data quality will be recorded in the database at the imaging session level and for each scan within the session. The database will include a binary pass/fail determination as well as fields for the aforementioned manual review criteria and the automated numeric QC metrics. Given the complexity and volume of image data being acquired in the HCP protocol, we anticipate that individual scans within each imaging visit will vary in quality. A single fMRI run, for example, might include an unacceptable level of motion, whereas other scans for that subject are acceptable in quality. In such cases, data re-acquisition is unlikely. The appropriate strategy for handling missing datasets will be dependent on exactly which data are absent.
Pipeline execution. The various processing streams described above are complex and computationally demanding. In order to ensure that they are run consistently and efficiently across all subjects, we will utilize XNAT’s pipeline service to execute and monitor the processing. XNAT’s pipeline approach uses XML documents to formally define the sequence of steps in a processing stream, including the executable, execution parameters, and input data. As a pipeline executes, the pipeline service monitors its execution and updates its status in the database. When a pipeline exits, notifications will be sent to HCP staff to review the results, following pipeline-specific QC procedures similar to those used to review the raw data. Pipelines that require short latency (such as those associated with initial QC) will be executed on the HCP cluster, while those that are more computationally demanding but less time sensitive will be executed on the HPCS.
Provenance. Given the complexity of the data analysis streams described above, it will be crucial to keep accurate track of the history of processing steps for each generated file. Provenance records will be generated at two levels. First, a record of the computational steps executed to generate an image or connectivity map will be embedded within a NIFTI header extension. This record will contain sufficient detail that the image could be regenerated from the included information. Second, higher level metadata, such as pipeline version and execution date, will be written into an XCEDE-formatted XML document (Gadde et al., 2011) and imported into ConnectomeDB. This information will be used to maintain database organization as pipelines develop over time.
Data-sharing. The majority of the data collected and stored by the HCP will be openly shared using the open-access model recommended by the Science Commons11. The only data that will be withheld from open access are those that could identify individual study participants, which will be made available only for group analyses submitted through ConnectomeDB. Data will be distributed in a rolling fashion through quarterly releases over the course of Phase 2. Data will be released in standard formats, including DICOM, NIFTI, GIFTI, and CIFTI.
Given the scope and scale of the datasets, our aim of open and rapid data-sharing represents a significant challenge. To address this challenge, the HCP will use a tiered distribution strategy (Figure 4). The first tier includes dynamic access to condensed representations of connectivity maps and related data. The second distribution tier will allow users to download bundled subsets of the data. These bundles will be configured to be of high scientific value while still being small enough to download within a reasonable time. A third tier will allow users to request a portable hard drive populated by a more extensive bundle of HCP data. Finally, users needing access to extremely large datasets that are impractical to distribute will be able to obtain direct access to the HPCS to execute their computing tasks. This raises issues of prioritization, cost recovery, and user qualification that have yet to be addressed.
Figure 4. HCP data distribution tiers.
Some of the data acquired by the HCP could potentially be used to identify the study participants. We will take several steps to mitigate this risk. As mentioned above, sensitive DICOM header fields will be redacted and facial features in the images will be obscured. Second, the precision of sensitive data fields will be reduced in the open-access data set, in some cases binning numeric fields into categories. Finally, we will develop web services that will enable users to submit group-wise analyses that would operate on sensitive genetic data without providing users with direct access to individual subject data. For example, users could request connectivity difference maps of subjects carrying the ApoE4 allele versus ApoE2/3. The resulting group-wise data would be scientifically useful while preventing individual subject exposure. This approach requires care to ensure that requested groups are of sufficient size and the number of overall queries is constrained to prevent computationally driven approaches from extracting individual subject information.
Visualization
The complexity and diversity of connectivity-related data types described above result in extensive visualization needs for the HCP. To address these needs, CWB, developed on top of Caret software (Van Essen et al., 2001)12 will include both browser and desktop versions. The browser-based version will allow users to quickly view data from ConnectomeDB, while the desktop version will allow users to carry out more demanding visualization and analysis steps on downloaded data.
Connectome Workbench
Connectome Workbench is based on Caret6, a prototype Java-based version of Caret, and will run on recent versions of Linux, Mac OS X, and Windows. It will use many standard Caret features for visualizing data on surfaces and volumes. This includes multiple viewing windows and many display options. Major visualization options will include (i) data overlaid on surfaces or volume slices in solid colors to display parcels and other regions of interest (ROIs), (ii) continuous scalar variables to display fMRI data, shape features, connectivity strengths, etc., each using an appropriate palette; (iii) contours projected to the surface to delineate boundaries of cortical areas and other ROIs, (iv) foci that represent centers of various ROIs projected to the surface; and (v) tractography data represented by needle-like representations of fiber orientations in each voxel.
A “connectivity selector” option will load functional and structural connectivity data from the appropriate connectivity matrix file (dense or parcellated) and display it on the user-selected surface and/or volume representations (e.g., as in Figure 2). Because dense connectivity files will be too large and slow to load in their entirety, connectivity data will be read in from disk by random access when the user requests a connectivity map for a particular brainordinate or patch of brainordinates. For functional connectivity data, it may be feasible to use the more compact time-series datasets and to calculate on the fly the correlation coefficients representing connectivity.
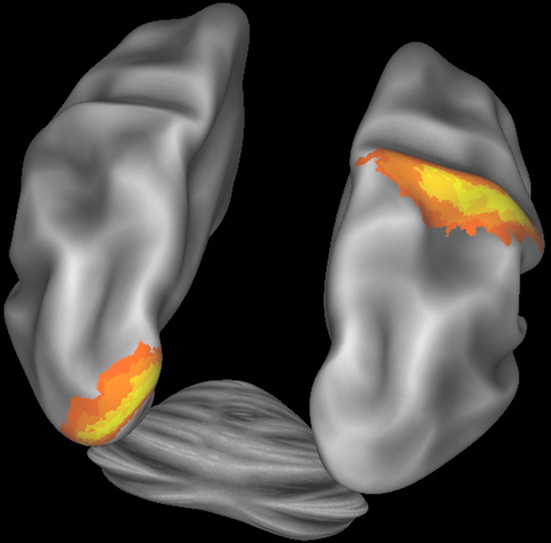
Figure 5 illustrates how CWB allows concurrent visualization of multiple brain structures (left and right cerebral hemispheres plus the cerebellum) in a single window. Subcortical structures will be viewable concurrently with surfaces or as volume slices in separate windows.
Figure 5. Connectome Workbench visualization of the inflated atlas surfaces for the left and right cerebral hemispheres plus the cerebellum. Probabilistic architectonic maps are shown of area 18 on the left hemisphere and area 2 on the right hemisphere.
Connectome Workbench will include options to display the results of various network analyses. For example, this may include concurrent visualization of network nodes in their 3D location in the brain as well as in a spring-embedded network, where node position reflects the strength and pattern of connectivity. The connection strength of graph edges will be represented using options of thresholding, color, and/or thickness. As additional methods are developed for displaying complex connectivity patterns among hundreds of nodes, the most useful of these will be incorporated either directly into CWB or via third party software.
Both the dense time-series and the parcellated time-series files provide temporal information related to brain activity. A visualization mode that plays “movies” by sequencing through and displaying each of the timepoints will be implemented. Options to view results of Task-fMRI paradigms will include both surface-based and volume-based visualization of individual and group-average data. Given that Task-fMRI time courses can vary significantly across regions (e.g., Nelson et al., 2010), options will also be available to view the average time course for any selected parcel or other ROI.
MEG and EEG data collected as part of the HCP will entail additional visualization requirements. This will include visualization in both sensor space (outside the skull) and after source localization to cortical parcels whose size respects the attainable spatial resolution. Representations of time course data will include results of power spectrum and BLP analyses.
ConnectomeDB/Workbench Integration
Querying ConnectomeDB from Connectome Workbench. While users will often analyze date already downloaded to their own computer, CWB will also be able to access data residing in the Connectome database. Interactions between the two systems will be enabled through ConnectomeDB’s web services API. CWB will include a search interface to identify subject groups in ConnectomeDB. Once a subject group has been selected, users can then visually explore average connectivity maps for this group by clicking on locations of interest on an atlas surface in CWB. With each click, a request to ConnectomeDB’s spatial query service will be submitted. Similar interactive explorations will be possible for all measures of interest, e.g., behavioral testing results or task performances from Task-fMRI sessions, with the possibility of displaying both functional and structural connectivity maps.
Browser-based visualization and Querying Connectome DB. Users will also be able to view connectivity patterns and other search results via the ConnectomeDB UI so that they can quickly visualize processed data without having to download data – and even view results on tablets and smart phones. To support this web-based visualization, we will develop a distributed CWB system in which the visualization component is implemented as a web-embeddable viewer using a combination of HTML5, JavaScript, and WebGL. The computational components of CWB will be deployed as a set of additional web services within the Connectome API. These workbench services will act as an intermediary between the viewer and ConnectomeDB, examining incoming visualization requests and converting them into queries on the data services API. Data retrieved from the database will then be processed as needed and sent to the viewer.
Links to external databases
Providing close links to other databases that contain extensive information about the human brain will further enhance the utility of HCP-related datasets. For example, the Allen Human Brain Atlas (AHBA)13 contains extensive data on gene expression patterns obtained by postmortem analyses of human brains coupled to a powerful and flexible web interface for data mining and visualization. The gene expression data(from microarray analyses and in situ hybridization analyses) have been mapped to the individual subject brains in stereotaxic space and also to cortical surface reconstructions. We plan to establish bi-directional spatially based links between CWB and the AHBA. This would enable a user of CWB interested in a particular ROI based on connectivity-related data to link to the AHBA and explore gene expression data related to the same ROI. Conversely, users of AHBA interested in a particular ROI based on gene expression data would be able to link to ConnectomeDB/Workbench and analyze connectivity patterns in the same ROI. A similar strategy will be useful for other resources, such as the SumsDB searchable database of stereotaxic coordinates from functional imaging studies14. Through the HCP’s outreach efforts, links to additional databases will be developed over the course of the project.
Discussion
By the end of Phase II, the WU-Minn HCP consortium anticipates having acquired an unparalleled neuroimaging dataset, linking functional, structural, behavioral, and genetic information in a large cohort of normal human subjects. The potential neuroscientific insights to be gained from this dataset are great, but in many ways unforeseeable. An overarching goal of the HCP informatics effort is to facilitate discovery by helping investigators formulate and test hypotheses by exploring the massive search space represented by its multi-modal data structure.
The HCP informatics approach aims to provide a platform that will allow for basic visualization of the dataset’s constituent parts, but will also encourage users to dynamically and efficiently make connections between the assembled data types. Users will be able to easily explore the population-average structural connectivity map, determine if the strength of a particular connection is correlated with a specific behavioral characteristic or genetic marker, or carry out a wide range of analogous queries. If the past decade’s experience in the domain of genome-related bioinformatics is a guide, data discovery is likely to take new and unexpected directions soon after large HCP datasets become available, spurring a new generation of neuroinformatics tools that are not yet imagined. We will be responsive to new methodologies when possible and will allow our interface to evolve as new discoveries emerge.
The HCP effort is ambitious in many respects. Its success in the long run will be assessed in many ways – by the number and impact of scientific publications drawing upon its data, by the utilization of tools and analysis approaches developed under its auspices, and by follow-up projects that explore brain connectivity in development, aging, and a myriad of brain disorders. From the informatics perspective, key issues will be whether HCP data are accessed widely and whether the tools are found to be suitably powerful and user-friendly. During Phase I, focus groups will be established to obtain suggestions and feedback on the many facets of the informatics platform and help ensure that the end product meets the needs of the target users. The outreach effort will also include booths and other presentations at major scientific meetings (OHBM, ISMRM, and SfN), webinars and tutorials, a regularly updated HCP website15, and publications such as the present one.
In addition to the open-access data that will be distributed by the HCP, the HCP informatics platform itself will be open source and freely available to the scientific community under a non-viral license. A variety of similar projects will likely emerge in the coming years that will benefit from its availability. We also anticipate working closely with the neuroinformatics community to make the HCP informatics system interoperable with the wide array of informatics tools that are available and under development.
While significant progress has been made since funding commenced for the HCP, many informatics challenges remain to be addressed. Many of the processing and analysis approaches to be used by the HCP are still under development and will undoubtedly evolve over the course of the project. How do we best handle the myriad of potential forks in processing streams? Can superseded pipelines be retired midway through the project or will users prefer for them to remain operational? What if a pipeline is found to be flawed? These and other data processing issues will require an active dialog with the user community over the course of the project. Subject privacy is another issue that requires both technical and ethical consideration. How do we minimize the risk of subject exposure while maximizing the utility of the data to the scientific community? Finally, what disruptive technologies may emerge over the 5 years of the HCP? How do we best maintain focus on our core deliverables while retaining agility to adopt important new tools that could further the scientific aims of the project? History suggests that breakthroughs can come from unlikely quarters. We anticipate that the HCP’s open data and software sharing will encourage such breakthroughs and contribute to the nascent field of connectome science and discovery.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Funded in part by the Human Connectome Project (1U54MH091657-01) from the 16 NIH Institutes and Centers that Support the NIH Blueprint for Neuroscience Research, by the McDonnell Center for Systems Neuroscience at Washington University, and by grant NCRR 1S10RR022984-01A1 for the CHCP; 1R01EB009352-01A1 and 1U24RR02573601 for XNAT support; and 2P30NS048056-06 for the NIAC. Members of the WU-Minn HCP Consortium are listed at http://www.humanconnectome.org/about/hcp-investigators.html and http://www.humanconnectome.org/about/hcp-colleagues.html. We thank Steve Petersen, Olaf Sporns, Jonathan Power, Andrew Heath, Deanna Barch, Jon Schindler, Donna Dierker, Avi Snyder, and Steve Smith for valuable comments and suggestions on the manuscript.
Footnotes
References
Beckmann, M., Johansen-Berg, H., and Rushworth, M. F. (2009). Connectivity-based parcellation of human cingulate cortex and its relation to functional specialization. J. Neurosci. 29, 1175–1190.
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text
de Pasquale, F., Della Penna, S., Snyder, A. Z., Lewis, C., Mantini, D., Marzetti, L., Belardinelli, P., Ciancetta, L., Pizzella, V., Romani, G. L., and Corbetta, M. (2010). Temporal dynamics of spontaneous MEG activity in brain networks. Proc. Natl. Acad. Sci. U.S.A. 107, 6040–6045.
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text
Feinberg, D. A., Moeller, S., Smith, S. M., Auerbach, E., Ramanna, S., Glasser, M. F., Miller, K. L., Ugurbil, K., and Yacoub, E. (2010). Multiplexed echo planar imaging for sub-second whole brain FMRI and fast diffusion imaging. PLoS ONE 5, e15710. doi: 10.1371/journal.pone.0015710
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text
Gadde, S., Aucoin, N., Grethe, J. S., Keator, D. B., Marcus, D. S., and Pieper, S. (2011). XCEDE: an extensible schema for biomedical data. _Neuroinformatics_1–14.
Gershon, R. C., Cella, D., Fox, N. A., Havlik, R. J., Hendrie, H. C., and Wagster, M. V. (2010). Assessment of neurological and behavioural function: the NIH Toolbox. Lancet Neurol. 9, 138–139.
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text
Jack, C. R. Jr., Bernstein, M. A., Fox, N. C., Thompson, P., Alexander, G., Harvey, D., Borowski, B., Britson, P. J., Whitwell, J. L., Ward, C., Dale, A. M., Felmlee, J. P., Gunter, J. L., Hill, D. L., Killiany, R., Schuff, N., Fox-Bosetti, S., Lin, C., Studholme, C., DeCarli, C. S., Krueger, G., Ward, H. A., Metzger, G. J., Scott, K. T., Mallozzi, R., Blezek, D., Levy, J., Debbins, J. P., Fleisher, A. S., Albert, M., Green, R., Bartzokis, G., Glover, G., Mugler, J., and Weiner, M. W. (2008). The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J. Magn. Reson. Imaging 27, 685–691.
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text
Johansen-Berg, H., and Behrens, T. E. (2009). From Quantitative Measurement in-vivo Neuroanatomy Boston, MA: Elsevier.
Marcus, D. S., Olsen, T. R., Ramaratnam, M., and Buckner, R. L. (2007). The Extensible Neuroimaging Archive Toolkit: an informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics 5, 11–34.
Pubmed Abstract | Pubmed Full Text
Nelson, S. M., Cohen, A. L., Power, J. D., Wig, G. S., Miezin, F. M., Wheeler, M. E., Velanova, K., Donaldson, D. I., Phillips, J. S., Schlaggar, B. L., and Petersen, S. E. (2010). A parcellation scheme for human left lateral parietal cortex. Neuron 67, 156–170.
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text
Newman, M. E. (2006). Modularity and community structure in networks. Proc. Natl. Acad. Sci. USA 103, 8577–8582.
Ou, W., Nummenmaa, A., Ahveninen, J., Belliveau, J. W., Hämäläinen, M. S., and Golland, P. (2010). Multimodal functional imaging using fMRI-informed regional EEG/MEG source estimation. NeuroImage 52, 97–108.
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text
Patel, V., Dinov, I. D., Van Horn, J. D., Thompson, P. M., and Toga, A. W. (2010). LONI MiND: metadata in NIfTI for DWI. Neuroimage 51, 665–676.
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text
Petrovic, V. S., Cootes, T. F., Mills, A. M., Twining, C. J., and Taylor, C. J. (2007). Automated analysis of deformable structure in groups of images. Proc. British Machine Vision Conference 2, 1060–1069.
Sabuncu, M. R., Singer, B. D., Conroy, B., Bryan, R. E., Ramadge, P. J., and Haxby, J. V. (2010). Function-based intersubject alignment of human cortical anatomy. Cereb. Cortex 20, 130–140.
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text
Scheeringa, R., Fries, P., Petersson, K.-M., Oostenveld, R., Grothe, I., Norris, D. G., Hagoort, P., and Bastiaansen, M. C. M. (2011). Neuronal dynamics underlying high- and low-frequency EEG oscillations contribute independently to the human BOLD signal. Neuron 69, 572–583.
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text
Sporns, O. (2010). Networks of the Brain. Cambridge, MA: MIT Press, 375 pp.
Van Essen, D. C., Drury, H. A., Dickson, J., Harwell, J., Hanlon, D., and Anderson, C. H. (2001). An integrated software suite for surface-based analyses of cerebral cortex. J. Am. Med. Inform. Assoc. 8, 443–459.
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text
Vincent, J. L., Patel, G. H., Fox, M. D., Snyder, A. Z., Baker, J. T., Van Essen, D. C., Zempel, J. M., Snyder, L. H., Corbetta, M., and Raichle, M. E. (2007). Intrinsic functional architecture in the anaesthetized monkey brain. Nature 447, 83−86.