“Megavirales”, a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses (original) (raw)
Abstract
The nucleocytoplasmic large DNA viruses (NCLDVs) comprise a monophyletic group of viruses that infect animals and diverse unicellular eukaryotes. The NCLDV group includes the families Poxviridae, Asfarviridae, Iridoviridae, Ascoviridae, Phycodnaviridae, Mimiviridae and the proposed family “Marseilleviridae”. The family Mimiviridae includes the largest known viruses, with genomes in excess of one megabase, whereas the genome size in the other NCLDV families varies from 100 to 400 kilobase pairs. Most of the NCLDVs replicate in the cytoplasm of infected cells, within so-called virus factories. The NCLDVs share a common ancient origin, as demonstrated by evolutionary reconstructions that trace approximately 50 genes encoding key proteins involved in viral replication and virion formation to the last common ancestor of all these viruses. Taken together, these characteristics lead us to propose assigning an official taxonomic rank to the NCLDVs as the order “Megavirales”, in reference to the large size of the virions and genomes of these viruses.
Access this article
Subscribe and save
- Get 10 units per month
- Download Article/Chapter or eBook
- 1 Unit = 1 Article or 1 Chapter
- Cancel anytime Subscribe now
Buy Now
Price excludes VAT (USA)
Tax calculation will be finalised during checkout.
Instant access to the full article PDF.
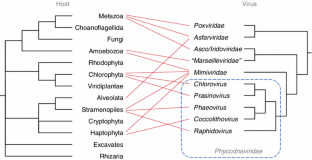
Fig. 1
Similar content being viewed by others
References
- Iyer LM, Aravind L, Koonin EV (2001) Common origin of four diverse families of large eukaryotic DNA viruses. J Virol 75:11720–11734. doi:10.1128/JVI.75.23.11720-11734.2001
Article PubMed CAS Google Scholar - Iyer LM, Balaji S, Koonin EV, Aravind L (2006) Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res 117:156–184. doi:10.1016/j.virusres.2006.01.009
Article PubMed CAS Google Scholar - Yutin N, Wolf YI, Raoult D, Koonin EV (2009) Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol J 6:223. doi:10.1186/1743-422X-6-223
Article PubMed Google Scholar - Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, La Scola B, Suzan M, Claverie JM (2004) The 1.2-megabase genome sequence of Mimivirus. Science 306:1344–1350. doi:10.1126/science.1101485
Article PubMed CAS Google Scholar - Claverie JM, Abergel C, Ogata H (2009) Mimivirus. Curr Top Microbiol Immunol 328:89–121
Article PubMed CAS Google Scholar - La Scola B, Campocasso A, N’Dong R, Fournous G, Barrassi L, Flaudrops C, Raoult D (2010) Tentative characterization of new environmental giant viruses by MALDI-TOF mass spectrometry. Intervirology 53:344–353. doi:10.1159/000312919
Article PubMed Google Scholar - Yoosuf N, Yutin N, Colson P, Shabalina SA, Pagnier I, Robert C, Azza S, Klose T, Wong J, Rossmann MG, La SB, Raoult D, Koonin EV (2012) Related giant viruses in distant locations and different habitats: Acanthamoeba polyphaga moumouvirus represents a third lineage of the Mimiviridae that is close to the megavirus lineage. Genome Biol Evol 4:1324–1330. doi:10.1093/gbe/evs109
Article PubMed CAS Google Scholar - Boyer M, Yutin N, Pagnier I, Barrassi L, Fournous G, Espinosa L, Robert C, Azza S, Sun S, Rossmann MG, Suzan-Monti M, La SB, Koonin EV, Raoult D (2009) Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc Natl Acad Sci USA 106:21848–21853. doi:10.1073/pnas.0911354106
Article PubMed CAS Google Scholar - Thomas V, Bertelli C, Collyn F, Casson N, Telenti A, Goesmann A, Croxatto A, Greub G (2011) Lausannevirus, a giant amoebal virus encoding histone doublets. Environ Microbiol 13:1454–1466. doi:10.1111/j.1462-2920.2011.02446.x
Article PubMed CAS Google Scholar - Colson P, Pagnier I, Yoosuf N, Fournous G, La Scola B, Raoult D (2012) Marseilleviridae, a new family of giant viruses infecting amoebae. Arch Virol 158(4):915–920. doi:10.1007/s00705-012-1537-y
Google Scholar - Claverie JM, Ogata H, Audic S, Abergel C, Suhre K, Fournier PE (2006) Mimivirus and the emerging concept of “giant” virus. Virus Res 117:133–144. doi:10.1016/j.virusres.2006.01.008
Article PubMed CAS Google Scholar - Arslan D, Legendre M, Seltzer V, Abergel C, Claverie JM (2011) Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae. Proc Natl Acad Sci USA 108:17486–17491
Article PubMed CAS Google Scholar - Fischer MG, Allen MJ, Wilson WH, Suttle CA (2010) Giant virus with a remarkable complement of genes infects marine zooplankton. Proc Natl Acad Sci USA 107:19508–19513. doi:10.1073/pnas.1007615107
Article PubMed CAS Google Scholar - Monier A, Claverie JM, Ogata H (2008) Taxonomic distribution of large DNA viruses in the sea. Genome Biol 9:R106. doi:10.1186/gb-2008-9-7-r106
Article PubMed Google Scholar - Monier A, Larsen JB, Sandaa RA, Bratbak G, Claverie JM, Ogata H (2008) Marine mimivirus relatives are probably large algal viruses. Virol J 5:12. doi:10.1186/1743-422X-5-12
Article PubMed Google Scholar - Yamada T (2011) Giant viruses in the environment: their origins and evolution. Curr Opin Virol 1:58–62
Article PubMed Google Scholar - Kristensen DM, Mushegian AR, Dolja VV, Koonin EV (2010) New dimensions of the virus world discovered through metagenomics. Trends Microbiol 18:11–19. doi:10.1016/j.tim.2009.11.003
Article PubMed CAS Google Scholar - Colson P, de Lamballerie X, Fournous G, Raoult D (2012) Reclassification of giant viruses composing a fourth domain of life in the new order Megavirales. Intervirology 55(5):321–332. doi:10.1159/000336562
Google Scholar - Yutin N, Koonin EV (2012) Hidden evolutionary complexity of nucleo-cytoplasmic large DNA viruses of eukaryotes. Virol J 9:161
Article PubMed CAS Google Scholar - Koonin EV, Yutin N (2010) Origin and evolution of eukaryotic large nucleo-cytoplasmic DNA viruses. Intervirology 53:284–292. doi:10.1159/000312913
Article PubMed Google Scholar - Krupovic M, Bamford DH (2008) Virus evolution: how far does the double beta-barrel viral lineage extend? Nat Rev Microbiol 6:941–948
Article PubMed CAS Google Scholar - Bahar MW, Graham SC, Stuart DI, Grimes JM (2011) Insights into the evolution of a complex virus from the crystal structure of vaccinia virus D13. Structure 19:1011–1020
Article PubMed CAS Google Scholar - Bigot Y, Asgari S, Bideshi D, Cheng XW, Federici BA et al. (2011) Family Ascoviridae. In: CM Fauquet, Mayo MA, Maniloff J, Desselberger U, Ball LA (eds) Virus taxonomy. Ninth report of the international committee on taxonomy of viruses, San Diego, pp 147–152
- Szajner P, Weisberg AS, Lebowitz J, Heuser J, Moss B (2005) External scaffold of spherical immature poxvirus particles is made of protein trimers, forming a honeycomb lattice. J Cell Biol 170:971–981
Article PubMed CAS Google Scholar - Seet BT, Johnston JB, Brunetti CR, Barrett JW, Everett H, Cameron C, Sypula J, Nazarian SH, Lucas A, McFadden G (2003) Poxviruses and immune evasion. Annu Rev Immunol 21:377–423
Article PubMed CAS Google Scholar - Werden SJ, Rahman MM, McFadden G (2008) Poxvirus host range genes. Adv Virus Res 71:135–171. doi:10.1016/S0065-3527(08)00003-1
Article PubMed CAS Google Scholar - Dixon LK, Abrams CC, Bowick G, Goatley LC, Kay-Jackson PC, Chapman D, Liverani E, Nix R, Silk R, Zhang F (2004) African swine fever virus proteins involved in evading host defence systems. Vet Immunol Immunopathol 100:117–134. doi:10.1016/j.vetimm.2004.04.002
Article PubMed CAS Google Scholar - Condit RC (2007) Vaccinia, Inc.—probing the functional substructure of poxviral replication factories. Cell Host Microbe 2:205–207
Article PubMed CAS Google Scholar - Mutsafi Y, Zauberman N, Sabanay I, Minsky A (2010) Vaccinia-like cytoplasmic replication of the giant Mimivirus. Proc Natl Acad Sci USA 107:5978–5982. doi:10.1073/pnas.0912737107
Article PubMed CAS Google Scholar - Netherton CL, Wileman TE (2013) African swine fever virus organelle rearrangements. Virus Res 173(1):76–86. doi:10.1016/j.virusres.2012.12.014
Google Scholar - Novoa RR, Calderita G, Arranz R, Fontana J, Granzow H, Risco C (2005) Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol Cell 97:147–172. doi:10.1042/BC20040058
Article PubMed CAS Google Scholar - Netherton CL, Wileman T (2011) Virus factories, double membrane vesicles and viroplasm generated in animal cells. Curr Opin Virol 1:381–387
Article PubMed CAS Google Scholar - Keeling PJ (2007) Genomics. Deep questions in the tree of life. Science 317:1875–1876
Article PubMed CAS Google Scholar - Keeling PJ, Burger G, Durnford DG, Lang BF, Lee RW, Pearlman RE, Roger AJ, Gray MW (2005) The tree of eukaryotes. Trends Ecol Evol 20:670–676
Article PubMed Google Scholar - Koonin EV (2010) The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol 11:209–211
Article PubMed Google Scholar
Acknowledgments
Natalya Yutin and Eugene V. Koonin are supported by intramural funds of the US Department of Health and Human Services (to the National Library of Medicine). James Van Etten is partially supported by NIH Grant P20 RR15635 from the COBRE Program of the National Center for Research Resources.
Conflict of interest
There are no potential conflicts of interest or financial disclosures for any of the authors.
Author information
Authors and Affiliations
- Unité des Rickettsies, URMITE UMR CNRS 7278 IRD 198 INSERM U1095, Facultés de Médecine et de Pharmacie, IHU Méditerranée Infection, Aix-Marseille Université, 27 Boulevard Jean Moulin, 13385, Marseille Cedex 05, France
Philippe Colson, Bernard La Scola & Didier Raoult - Pôle des Maladies Infectieuses et Tropicales Clinique et Biologique, Fédération de Bactériologie-Hygiène-Virologie, IHU Méditerranée Infection, Assistance Publique, Hôpitaux de Marseille, Centre Hospitalo-Universitaire Timone, 264 Rue Saint-Pierre, 13385, Marseille Cedex 05, France
Philippe Colson, Xavier De Lamballerie, Bernard La Scola & Didier Raoult - Unité des Virus Emergents, UMR190 Emergence des pathologies virales, Faculté de Médecine, Institut de Recherche pour le Développement, EHSP French School of Public Health, Aix-Marseille Université, Marseille, France
Xavier De Lamballerie - National Center for Biotechnology Information (NCBI), National Library of Medicine, National Institutes of Health, Bldg. 38A, 8600 Rockville Pike, Bethesda, MD, 20894, USA
Natalya Yutin & Eugene V. Koonin - School of Biological Sciences, The University of Queensland, St Lucia, QLD, 4072, Australia
Sassan Asgari - UMR INRA-CNRS 7247, PRC, Centre INRA de Nouzilly, 37380, Nouzilly, France
Yves Bigot - Molecular and Developmental Biology, University of California, Riverside, CA, 92521, USA
Dennis K. Bideshi - Department of Microbiology, Miami University, Oxford, OH, 45056, USA
Xiao-Wen Cheng - Department of Entomology, University of California, Riverside, CA, 92521, USA
Brian A. Federici - Department of Plant Pathology, University of Nebraska, Lincoln, NE, 68583-0722, USA
James L. Van Etten - Nebraska Center for Virology, University of Nebraska, Lincoln, NE, 68583-0900, USA
James L. Van Etten
Authors
- Philippe Colson
You can also search for this author inPubMed Google Scholar - Xavier De Lamballerie
You can also search for this author inPubMed Google Scholar - Natalya Yutin
You can also search for this author inPubMed Google Scholar - Sassan Asgari
You can also search for this author inPubMed Google Scholar - Yves Bigot
You can also search for this author inPubMed Google Scholar - Dennis K. Bideshi
You can also search for this author inPubMed Google Scholar - Xiao-Wen Cheng
You can also search for this author inPubMed Google Scholar - Brian A. Federici
You can also search for this author inPubMed Google Scholar - James L. Van Etten
You can also search for this author inPubMed Google Scholar - Eugene V. Koonin
You can also search for this author inPubMed Google Scholar - Bernard La Scola
You can also search for this author inPubMed Google Scholar - Didier Raoult
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toDidier Raoult.
Additional information
S. Asgari, Y. Bigot, D. K. Bideshi, X.-W. Cheng and B. A. Federici belong to the Ascoviridae study group of the International Committee on Taxonomy of Viruses (ICTV).
Rights and permissions
About this article
Cite this article
Colson, P., De Lamballerie, X., Yutin, N. et al. “Megavirales”, a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses.Arch Virol 158, 2517–2521 (2013). https://doi.org/10.1007/s00705-013-1768-6
- Received: 03 August 2012
- Accepted: 07 May 2013
- Published: 29 June 2013
- Issue Date: December 2013
- DOI: https://doi.org/10.1007/s00705-013-1768-6