Genetic variation of Ginkgo biloba L. (Ginkgoaceae) based on cpDNA PCR-RFLPs: inference of glacial refugia (original) (raw)
Introduction
The distribution and genetic diversity of plant species have been deeply modified by Pleistocene glaciations (Comes and Kadereit, 1998; Hewitt, 1999, 2004; Newton et al, 1999; Abbott and Brochmann, 2003). During glaciations, the ranges of species changed and some ancestral populations became extinct, and recolonization subsequently occurred during interglacial periods (Widmer and Lexer, 2001). The species genomes were dramatically influenced by these processes, and molecular markers can be used to infer the phylogeographic pattern of species in response to the Pleistocene climatic changes (Newton et al, 1999; Shen et al, 2002; Abbott and Comes, 2004).
In contrast to intense investigations on plants of Europe and northern America (Hewitt, 2000; Shen et al, 2002), understanding of the effects of past climatic events on population structure and phylogeny of species in Asia is relatively limited (Szmidt and Wang, 1993; Wang and Szmidt, 1994; Szmidt et al, 1996; Chen et al, 1997; Tomaru et al, 1997; Lin, 2001; Huang et al, 2002; Lu et al, 2002; Okaura and Harada, 2002; Aoki et al, 2004). The Eurasian ice sheets covered millions of square kilometers of the continent in the late Pleistocene period (Svendsen et al, 1999). It is accepted that, during the Quaternary, mountains over 3500 m in western China became glaciated, but there is disagreement over whether the lower central and eastern mountains (<3000 m) were glaciated (Shi et al, 1989). Temperatures are estimated to have been 5–13°C lower than at present in China (Shi et al, 1989): low enough to have profound impact on plants (Wang and Liu, 1994).
Ginkgos (Ginkgoaceae) have a history dating back approximately to the early Permian (∼280 MA) (Willis and McElwain, 2002). At the height of their global radiation, fossil evidence indicates that there were at least 16 genera and that they formed a significant part of the world vegetation (Willis and McElwain, 2002). Today, they are represented by a single species, Ginkgo biloba, which is endemic to China. This species was listed as a rare species in the 1997 IUCN red list of threatened plants (http://www.unep-wcmc.org/) and listed in the red list of endangered plant species of China (Fu and Jin, 1992). Thus, it should receive effective conservation. However, the existence of natural G. biloba populations is disputed and the refugia during Pleistocene glaciations are not known (Liang and Li, 2001). The West Tianmu Mountain of Zhejiang Province is one likely refugium (Liang and Li, 2001; Lin and Zhang, 2004). Recently, several papers have suggested that some other sites had native populations, based on plant community surveys and human settlement history (Zhou et al, 1982; Xiang and Xiang, 1997; Xiang et al, 1998; Li et al, 1999), although this evidence might be considered inconclusive.
Molecular markers have proved to be useful tools for identifying refugia and tracking postglacial migration routes of plant species. Chloroplast DNA (cpDNA) is maternally inherited in most flowering plants and some gymnosperms (such as Ginkgo, Cycas and Ephedra) (Mogensen, 1996). It therefore provides a seed-specific marker. Maternally inherited markers tend to display highly structured geographical distribution because of the lower potential for dispersal by seeds than by pollen (Newton et al, 1999). It is consequently easier to elucidate the refugia and migration history of plant species using maternally inherited markers (rather than biparently inherited ones) (Newton et al, 1999). In the present paper, we try to understand the cpDNA haplotype diversity of G. biloba, and to infer the potential refugia.
Materials and methods
Species and sampling
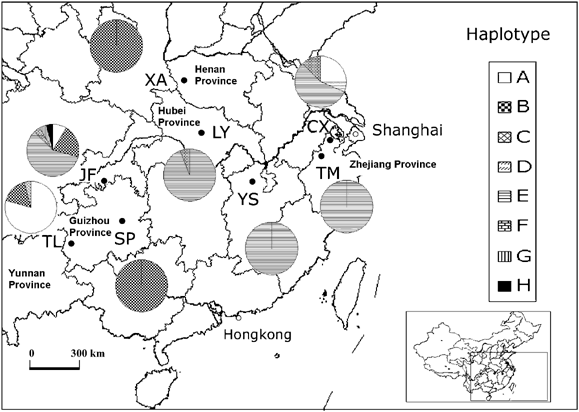
G. biloba is a long-lived deciduous species. It occurs through most of mainland China and many other countries. However, most trees have been planted for ornamental or medical purposes. Several sites were reported to have natural Ginkgo based on plant community surveys or/and the history of human settlement (Table 1). We will call them ‘natural’ populations because no convincing evidence exists to the contrary. We sampled populations covering these sites (six populations) as well as two additional planted populations (Table 1, Figure 1). They can be divided into three regions: southwestern China (JF, TL and SP), eastern China (TM, CX and YS) and central China (LY and XA).
Table 1 Location and sample size of G. biloba populations in the present study
Figure 1
Geographic locations of eight populations of G. biloba in China. TL: Tuole of Pan County, Guizhou Province; CX: Changxing County, Zhejiang Province; LY: Luoyang of Suizhou City, Hubei Province; YS: Yushan of Yongxiu County, Jiangxi Province; SP: Shaping of Guiyang City, Guizhou Province; TM: West Tianmu Mountain of Linan City, Zhejiang Province; XA: Xiaan of Xixia County, He’nan Province; JF: Jinfou Mountain of Nanchuan County, Chongqing Municipality.
Buds were collected from trees in West Tianmu Mountain (TM) and brought fresh to the laboratory for DNA extraction, while leaves were collected from individuals of the other seven populations and dried using silica gel has described previously (Chase and Hills, 1991). In all, 11–27 trees were sampled from each location. In order to avoid sampling recently planted trees, we sampled those with a DBH (diameter at breast-height) as large as possible. The DBH of the sampled trees ranged from 39 to 210 cm, of which only 10 (three in SP and TM and four in XA population) were less than 50 cm (Table 1).
DNA extraction
Total DNA was extracted using a miniprep CTAB protocol modified from Doyle and Doyle (1987). Dry leaves were ground to powder with a cold grinder, suspended in 750 μl 60°C CTAB 2 × buffer and incubated for 90 min. After chloroform isoamyl extraction, the aqueous phase was collected and digested with RNase A for 30 min. Finally, the nucleic acid was precipitated with 100% ethanol and resuspended in TE buffer.
PCR-RFLP of cpDNA
PCR assays were performed in 50 μl total volume containing 0.2 μM of each primer, 10 mM KCl, 8 mM (NH4)2SO4, 10 mM Tris-HCl (pH 9.0), 2.5 mM MgCl2, 200 μM dNTPs, 2 U of Taq DNA polymerase and 40 ng template DNA. In total, 16 pairs of universal primers for noncoding regions of cpDNA (Taberlet et al, 1991; Demesure et al, 1995; Dumolin-Lapegue et al, 1997b) were used in initial screening. No product was obtained using primers for _trnf_M-_psa_A, _trn_V-_rbc_L and _trn_F-_trn_V (Dumolin-Lapegue et al, 1997b).
Fragments of _trn_H-_trn_K, _trn_C-_trn_D, _psa_A-_trn_S, _trn_S-_trn_T, _trn_K-_trn_Q, _trn_Q-_trn_R, _rpo_C-_trn_C, _trn_T-_psb_C and _trn_T-_trn_F were of low quality and not suitable for RFLP analysis. The other four pairs of primers for _trn_K1-_trn_K2, _trn_D-_trn_T, _psb_C-_trn_S and _trn_S-_trnf_M produced fragments of suitable quality, which then were digested with the endonucleases _Rsa_I, _Hinf_I and _Hae_III, respectively. After digestion at 37°C for 6 h, the products were resolved electrophoretically on either 2% agarose gels or 8% polyacrylamide gels stained with ethidium bromide and photographed using Bio-Rad Gel Doc2000.
Data analysis
Diversity measures were calculated directly on the haplotype frequencies using Lewis and Zaykin's (2000) GDA program (version 1.0 d15). The number of haplotypes and haplotype diversity (ĥ) were calculated for each population and at region and species level, according to Nei (1987). Gene flow among populations was characterized as Nm, the estimated number of female migrants per generation between populations, using the formula F_ST=1/(1+2_Nm) where N is the female effective population size and m is the female migration rate.
Results
With the enzymes _Rsa_I, _Hinf_I and _Hae_III, no variation was observed in restriction patterns of the amplification products of primers for _trn_D-_trn_T, _psb_C-_trn_S and _trn_S-_trn_fM. Variants were observed for _trn_K1-_trn_K2 products when digested by _Rsa_I, _Hinf_I and _Hae_III (Table 2). All the variants are caused by point mutations. Eight haplotypes were defined (A_–_H). Three were common, with frequencies of 0.474 (haplotype E), 0.314 (haplotype B) and 0.160 (haplotype A), four were rare (frequencies equal to 0.006 for haplotype C, D, G and H), and haplotype F was infrequent, with a frequency of 0.026.
Table 2 Haplotype definitions, with the restriction profiles of _trn_K1-_trn_K2 by each endonuclease, and measures of differentiation (θ) in the study populations of G. biloba
Population JF has the highest effective number of haplotypes (2.53), followed by CX (2.51) and TL (1.52). Similar trends are observed for the actual numbers of haplotypes (Table 3). Haplotype diversity (ĥ) is greatest in population CX, followed by JF and TL (Table 3). One other population (LY), almost fixed for haplotype E, has one additional variant (_ĥ_=0.121), and the remaining four population samples are fixed for one haplotype. The mean ĥ is 0.214 and ĥ at the species level is 0.652. At the regional level, there are 7, 4 and 3 haplotypes for southwestern, eastern and central China, respectively. The value of ĥ ranges from 0.246 (eastern China) to 0.685 (southwestern China) with a mean of 0.485. The overall value of θ is 0.676 (Walter and Epperson, 2001), and the estimate of Nm is 0.240.
Table 3 Frequencies and diversity of haplotypes within study populations of G. biloba
Nei’s (1978) genetic distance reflects the same pattern of haplotype differentiation between pairs of populations (Table 4).
Table 4 Genetic distance a based on haplotype frequencies
Discussion
cpDNA polymorphism in G. biloba
With three different restriction enzymes, eight different cpDNA haplotypes were found in 158 individuals (Table 2). The level of genetic variation is moderate compared to that of other plants, for example, three haplotypes in 52 Silene hifacensis individuals (Prentice et al, 2003), 10 cpDNA haplotypes in 188 Beta vulgaris ssp. maritima individuals (Forcioli et al, 1998), 13 haplotypes in 217 Alnus glutinosa individuals (King and Ferris, 1998) and 23 haplotypes in 1412 white oaks (Dumolin-Lapegue et al, 1997a).
At the regional level, southeastern China has the highest haplotype diversity and central China the lowest. Genetic variation at the population level is much more variable. Population JF has as many as six haplotypes, while the other four populations have only one haplotype. It is intriguing that CX has more haplotypes than some other sites and has the highest haplotype diversity (0.621). The CX population is believed to have been transplanted from other sites, hence artificial admixture can explain the high diversity, if introduced individuals were drawn from several populations (Wade and McCauley, 1988). High genetic diversity in nonrefugia population has been observed in other plant species. Pinus resinosa, for example, shows the highest diversity in previously glaciated areas (Walter and Epperson, 2001) due to the natural postglacial recolonization from different refugia.
It has been shown theoretically and empirically that the level of genetic differentiation among populations is expected to be higher for maternally inherited cpDNA markers than for biparentally inherited nuclear genes (Ennos, 1994; Raspe et al, 2000). In accordance with this expectation, a _G_ST of 0.161 was observed for RAPD markers in G. biloba (Fan et al, 2004), whereas a much higher level of differentiation (_θ_=0.676) was found for cpDNA haplotypes in the present study. Limited gene flow and drift may contribute to the difference (Raspe et al, 2000).
Glacial refugia of G. biloba
The earliest known fossil of Ginkgo is from the Middle Jurassic epoch of 170 Myr ago and evolutionary history became clear with a recent find (Zhou and Zheng, 2003). East Asia provided the later habitats of Ginkgo species, and fossils have been found from the Oligocene, Miocene, Pliocene and early Pleistocene in Japan (Uemura, 1997). It has been suggested that West Tianmu Mountain was the refuge of G. biloba during the last glaciations, although there has been some disagreement (Liang and Li, 2001; Lin and Zhang, 2004) due to lack of a detailed fossil record (Zhou, 2003).
The cpDNA variation reported here indicates strong genetic differentiation and shows that the spread of G. biloba since the last glaciation does not coincide with previous expectations. The pattern suggests that southwestern China was a refugium for G. biloba. Seven of the eight haplotypes were found there and three of them were endemic to this region.
It would not be too surprising if the refugium of G. biloba was located in SW China during the Pleistocene ice ages. SW China is located southeast of the Tibetan Plateau and south to the Qing Mountains. The plateau and mountains form a natural division between the temperate zone and subtropical zone in mainland China. Consequently, SW China was less affected by cold air from Siberia during the glaciations. This region has the greatest biodiversity in China due to the relatively stable environment and diverse topography. It is also one of the 25 global biodiversity hotspots (Myers et al, 2000). The Yunnan Province, for example, has been estimated to harbor more than 15 000 vascular plant species. Many other living fossils are also found there. For example, Cathaya argyrophylla, Tetracentron sinense and natural Metasequoia glyptostroboides populations have been found in remote mountains of this region. Wang and Liu (1994) also suggested that this region is an important refugium for species surviving the Pleistocene glaciations.
The most likely refugium is sited in Jinfoshan, at the boundary of Chonqing Municiplity and Guizhou Province, as suggested by the current occurrence of ancient Ginkgo trees. Recently, a natural-like Ginkgo forest was reported in Jinfoshan by Li et al (1999) where there are 70 trees with DBH larger than 1 m and eight with DBH larger than 2 m. The town was established about 200 years ago and the largest Ginkgo tree was estimated to be 2500 years old with a DBH of 3.69 m (Li et al, 1999). Wuchuan County of Guizhou Province, not far from Jinfoshan, was also reported to have a natural Ginkgo population (Xiang and Xiang, 1997; Xiang et al, 1998). We sampled and analyzed the Wuchuan population. However, for unknown reasons, no amplified product was obtained with the universal primers.
Population TL, located around the border of Guizhou Province and Yunnan Province, seems to be another potential refugium because a higher than average number of haplotypes was found there and one of the haplotypes is only found there. The historical record of human colonization also supports this conclusion. It has been reported that there were 918 ancient Ginkgo trees in and around Tuole Village of Pan County (Xiang et al, 2003). The oldest is older than 1000 years, whereas the village was established about 500 years ago (Xiang et al, 2003).
Population SP, not far from JF and TL, is fixed for haplotype B that is not the most frequence haplotype in JF and TL. This may reflect loss of genetic variation due to a bottleneck during glaciation. Similarly, Ginkgo in other natural populations that may have become established after the retreat of the glaciers and bottlenecks during range expansion may explain the low haplotype diversity in these populations (Widmer and Lexer, 2001).
There are many papers discussing whether G. biloba population in Tianmu Mountain is natural or transplanted (Hu, 1954; Liang and Li, 2001; Lin and Zhang, 2004). In 1954, Hu, who described the living fossil Metasequoia glyptostroboides, reported there were some wild individuals in Zhejiang besides the planted ones (Hu, 1954). Since then, several researchers have proposed that West Tianmu Mountain was the sole refugium of G. biloba (Lin and Zhang, 2004), while others disagreed (Liang and Li, 2001). The present study indicates that West Tianmu Mountain does not seem to have served as a refugium for _Ginkgo_s since there is only one common cpDNA haplotype. This conclusion is in agreement with the settlement history. West Tianmu Mountain is famous for temples. The oldest Old Temple, was built in 936 AD The largest ginkgo tree is 123 cm in DBH and estimated to be about 500 years old. Hence, the Old Temple was built much earlier than the largest ginkgo tree. Of the many tree species planted around temples, the Buddhist monks preferred Ginkgo. As a result, there are many large ginkgo trees in or around temples in China, with some more than 1000 years old. Therefore, ginkgos in West Tianmu Mountain are likely to have been planted by monks, and may not be natural.
References
- Abbott RJ, Brochmann C (2003). History and evolution of the arctic flora: in the footsteps of Eric Hulten. Mol Ecol 12: 299–313.
Article PubMed Google Scholar - Abbott RJ, Comes HP (2004). Evolution in the Arctic: a phylogeographic analysis of the circumarctic plant, Saxifraga oppositifolia (Purple saxifrage). New Phytol 161: 211–224.
Article CAS Google Scholar - Aoki K, Suzuki T, Hsu T-W, Murakami N (2004). Phylogeography of the component species of broad-leaved evergreen forests in Japan, based on chloroplast DNA variation. J Plant Res 117: 77–94.
Article PubMed Google Scholar - Chase MW, Hills HH (1991). Silica gel: an ideal material for field preservation of leaf samples for DNA studies. Taxon 40: 215–220.
Article Google Scholar - Chen XY, Wang XH, Song YC (1997). Genetic diversity and differentiation of Cyclobalanopsis glauca populations in East China. Acta Bot Sin 39: 149–155.
Google Scholar - Comes HP, Kadereit JW (1998). The effect of quaternary climatic changes on plant distribution and evolution. Trends Plant Sci 3: 432–438.
Article Google Scholar - Demesure B, Sodzi N, Petit RJ (1995). A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Mol Ecol 4: 129–131.
Article CAS PubMed Google Scholar - Doyle JJ, Doyle JL (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19: 11–15.
Google Scholar - Dumolin-Lapegue S, Demesure B, Fineschi S, Corre VL, Petit RJ (1997a). Phylogeographic structure of white oaks throughout the European continent. Genetics 146: 1475–1487.
CAS PubMed PubMed Central Google Scholar - Dumolin-Lapegue S, Pemonge M-H, Petit RJ (1997b). An enlarged set of consensus primers for the study of organelle DNA in plants. Mol Ecol 6: 393–398.
Article CAS PubMed Google Scholar - Ennos RA (1994). Estimating the relative rates of pollen and seed migration among plants populations. Heredity 72: 250–259.
Article Google Scholar - Fan XX, Shen L, Zhang X, Chen XY, Fu CX (2004). Assessing genetic diversity of Ginkgo biloba L. (Ginkgoaceae) populations from China by RAPD markers. Biochem Genet 42: 269–278.
Article CAS PubMed Google Scholar - Forcioli D, Saumitou-Laprade P, Valero M, Vernet P, Cuguen J (1998). Distribution of chloroplast DNA diversity within and among populations in gynodioecious Beta vulgaris ssp. maritima (Chenopodiaceae). Mol Ecol 7: 1193–1204.
Article CAS Google Scholar - Fu LG, Jin JM (1992). Red List of Endangered Plants in China. Science Press: Beijing.
Google Scholar - Hewitt G (2000). The genetic legacy of the Quaternary ice ages. Nature 405: 907–913.
Article CAS PubMed Google Scholar - Hewitt GM (1999). Post-glacial re-colonization of European biota. Biol J Linn Soc 68: 87–112.
Article Google Scholar - Hewitt GM (2004). Genetic consequences of climatic oscillations in the Quaternay. Phil Trans Roy Soc Lond B 359: 183–195.
Article CAS Google Scholar - Hu X-S (1954). Metasequoia glyptostroboides, Glyptostrobus pensilis and Ginkgo biloba. Biol Bullet Sin 12: 12–15.
- Huang SSF, Hwang S-Y, Lin T-P (2002). Spatial pattern of chloroplast DNA variation of Cyclobalanopsis glauca in Taiwan and East Asia. Mol Ecol 11: 2349–2358.
Article CAS PubMed Google Scholar - King RA, Ferris C (1998). Chloroplast DNA phylogeography of Alnus glutinosa (L.) Gaertn. Mol Ecol 7: 1151–1161.
Article CAS Google Scholar - Li JW, Liu ZY, Tan YM, Ren MB (1999). Studies on the Ginkgo at the Jinfoshan Mountain. For Res 12: 197–201.
CAS Google Scholar - Liang LX, Li SN (2001). Argument about wild populations of Ginkgo. Sci Silvae Sin 37: 135–137.
Google Scholar - Lin H, Zhang DH (2004). Analysis for the origin of Ginkgo population in Tianmu Mountain. Sci Silvae Sin 40: 28–31.
Google Scholar - Lin T-P (2001). Allozyme variations in Michelia formosana (Kanehira) Masamune (Magnoliaceae), and the inference of a glacial refugium in Taiwan. Theor Appl Genet 102: 450–457.
Article CAS Google Scholar - Lu S-Y, Hong K-H, Liu S-L, Cheng Y-P, Wu W-L, Chiang T-Y (2002). Genetic variation and population differentiation of Michelia formosana (Magnoliaceae) based on cpDNA variation and RAPD fingerprints: relevance to post-Pleistocene recolonization. J Plant Res 115: 203–216.
Article CAS PubMed Google Scholar - Mogensen HL (1996). The hows and whys of cytoplasmic inheritance in seed plants. Am J Bot 83: 383–404.
Article Google Scholar - Myers N, Mittermeier RA, Mittermeier CG, Fonseca GAB, Kent J (2000). Biodiversity hotspots for conservation priorities. Nature 403: 853–858.
Article CAS PubMed Google Scholar - Nei M (1987). Molecular Evolutionary Genetics. Columbia University Press: New York.
Google Scholar - Newton AC, Allnutt TR, Gillies ACM, Lowe AJ, Ennos RA (1999). Molecular phylogeography, intraspecific variation and the conservation of tree species. Trends Ecol Evol 14: 140–145.
Article CAS PubMed Google Scholar - Okaura T, Harada K (2002). Phylogeographical structure revealed by chloroplast DNA variation in Japanese beech (Fagus crenata Blume). Heredity 88: 322–329.
Article CAS PubMed Google Scholar - Prentice HC, Malm JU, Mateu-Andres I, Segarra-Moragues JG (2003). Allozyme and chloroplast DNA variation in island and mainland populations of the rare Spanish endemic, Silene hifacensis (Caryophyllaceae). Conserv Genet 4: 543–555.
Article CAS Google Scholar - Raspe O, Saumitou-Laprade P, Cuguen J, Jacquemart A-L (2000). Chloroplast DNA haplotype variation and population differentiation in Sorbus aucuparia L. (Rosaceae: Maloideae). Mol Ecol 9: 1113–1122.
Article CAS PubMed Google Scholar - Shen L, Chen XY, Li YY (2002). Glacial refugia and postglacial recolonization patterns of organisms. Acta Ecol Sin 22: 1983–1990.
Google Scholar - Shi YF, Cui ZJ, Li JJ (1989). Quaternay Glacials and Environmental Issues of Eastern China. Science Press: Beijing.
Google Scholar - Svendsen JI, Astakhov VI, Bolshiyanov DYU (1999). Maximum extent of the Eurasian ice sheets in the Barents and Kara Sea region during the Weichselian. Boreas 28: 234–242.
Article Google Scholar - Szmidt AE, Wang XR (1993). Molecular systematics and genetic differentiation of Pinus sylvestris (L.) and P. densiflora (Sieb. et Zucc.). Theor Appl Genet 86: 159–165.
Article CAS PubMed Google Scholar - Szmidt AE, Wang XR, Changtragoon S (1996). Contrasting patterns of genetic diversity in two tropical pines: Pinus kesiya (Royle ex Gordon) and P. merkusii (Jungh et De Vriese). Theor Appl Genet 92: 436–441.
Article CAS PubMed Google Scholar - Taberlet P, Gielly L, Pautou G, Bouvet J (1991). Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol 17: 1105–1109.
Article CAS PubMed Google Scholar - Tomaru N, Mitsutsuji T, Takahashi M, Tsumura Y, Uchida K, Ohba K (1997). Genetic diversity in Fagus crenata (Japanese beech): influence of the distributional shift during the late-Quaternary. Heredity 78: 241–251.
Article Google Scholar - Uemura K (1997). Cenozoic history of Ginkgo in East Asia. In: Hori T, Ridge RW, Tulecke W, Tredici PD, Tremouillaux-Guiller J Tobe H (eds) Ginkgo biloba – A Global Treasure from Biology to Medicine. Springer Verlag: Tokyo, pp 207–221.
Chapter Google Scholar - Wade MJ, McCauley DE (1988). Extinction and colonization: their effects on the genetic differentiation of local population. Evolution 42: 995–1005.
Article PubMed Google Scholar - Walter R, Epperson BK (2001). Geographic pattern of genetic variation in Pinus resinosa: area of greatest diversity is not the origin of postglacial populations. Mol Ecol 10: 103–111.
Article CAS PubMed Google Scholar - Wang XP, Liu YK (1994). Theory and Practice of Biodiversity. China Environmental Science Press: Beijing.
Google Scholar - Wang XR, Szmidt AE (1994). Hybridisation and chloroplast DNA variation in a Pinus species complex from Asia. Evolution 48: 1020–1031.
Article CAS PubMed Google Scholar - Widmer A, Lexer C (2001). Glacial refugia: sanctuaries for allelic richness, but not for gene diversity. Trends Ecol Evol 16: 267–268.
Article CAS PubMed Google Scholar - Willis KJ, McElwain JC (2002). The Evolution of Plants. Oxford University Press: New York.
Google Scholar - Xiang H, Tu CL, Xiang YH (2003). A report on Ginkgo resources in Panxian County, Guizhou Province: Guizhou ancient Ginkgo resources investigation 5. Guizhou Sci 21: 159–174.
Google Scholar - Xiang YH, Lu XC, Xiang BX (1998). Ancient Ginkgo biloba report II: data of ancient Ginkgo biloba deleted communties in Luping Village and Fengle Town of Wuchuan County Guizhou Province. Guizhou Sci 16: 241–252.
Google Scholar - Xiang YH, Xiang BX (1997). Primary report on an ancient Ginkgo biloba deleted community in Wuchuan County, Guizhou Province. Guizhou Sci 15: 239–244.
Google Scholar - Xiang YH, Xiang Z (1999). Ancient _Ginkgo bi_loba report III. investigation on ancient Ginkgo biloba remanant population in Guiyang. Guizhou Sci 17: 221–230.
Google Scholar - Zhou YL, Bao J, Huang HQ (1982). Research of the gingko's primitive organisms’ community in Suizhou Forestry Bureau. Hubei For SciTech 104: 35–38.
Google Scholar - Zhou Z, Zheng S (2003). Missing link in Ginkgo evolution. Nature 423: 821–822.
Article CAS PubMed Google Scholar - Zhou ZY (2003). Mesozoic Ginkgoaleans: phyogeny, classification and evolutionary trends. Acta Bot Yunnanica 25: 377–396.
Google Scholar
Acknowledgements
We thank Mrs Tianyi Wu of Biology Department of Marqueet University for reading and providing helpful comments on the manuscript and Mrs Shuzhen Yang of Bureau of Tianmu Mountain National Nature Reserve for providing help in collecting samples. We acknowledge two anonymous reviewers for criticial comments on an early edition of the manuscript and Dr David Rosenthal for improving the language. We also thank Professor Ying JS of Institute of Botany and Professor Jiang MX of Wuhan Institute of Botany for providing helpful information about G. biloba. The work was supported in part by the State Key Basic Research and Development Plan of China (G2000046806), Shuguang Scholar Program and ‘211’ Program to X-Y Chen.
Author information
Author notes
- L Shen
Present address: Department of Plant Biology, The University of Georgia, Athens, GA 30602, USA
Authors and Affiliations
- Department of Environmental Sciences, East China Normal University, Shanghai, 200062, China
L Shen, X-Y Chen, X Zhang & Y-Y Li - Lab of Plant Systematic Evolution and Biodiversity, College of life Science, Zhejiang University, Hangzhou, 310029, China
C-X Fu & Y-X Qiu
Authors
- L Shen
You can also search for this author inPubMed Google Scholar - X-Y Chen
You can also search for this author inPubMed Google Scholar - X Zhang
You can also search for this author inPubMed Google Scholar - Y-Y Li
You can also search for this author inPubMed Google Scholar - C-X Fu
You can also search for this author inPubMed Google Scholar - Y-X Qiu
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toX-Y Chen.
Rights and permissions
About this article
Cite this article
Shen, L., Chen, XY., Zhang, X. et al. Genetic variation of Ginkgo biloba L. (Ginkgoaceae) based on cpDNA PCR-RFLPs: inference of glacial refugia.Heredity 94, 396–401 (2005). https://doi.org/10.1038/sj.hdy.6800616
- Received: 15 September 2004
- Accepted: 23 September 2004
- Published: 10 November 2004
- Issue Date: 01 April 2005
- DOI: https://doi.org/10.1038/sj.hdy.6800616