GitHub - lgatto/pRolocGUI: Interactive visualisation and exploration of spatial proteomics data (original) (raw)
Bioconductor build status:
Exploring and visualising spatial proteomics data
Introduction
ThepRolocGUIpackage is an interactive interface to explore and visualise experimental mass spectrometry-based spatial proteomics data. It relies on the shiny framework for interactive visualisation, theMSnbasepackage to handle data and metadata and thepRolocsoftware for spatial proteomics specific data matters. Example spatial data is available in thepRolocdataexperiment package.
The pRoloc suite set of software are distributed as part of theR/Bioconductor project and are developed by Lisa Breckels at the Cambridge Centre for Proteomicsat the University of Cambridge and by Laurent Gatto, director of the Computational Biology and Bioinformatics (CBIO) group at UCLouvain, in Belgium.
This document describes the installation of the software, followed by a basic quick start guide for using pRolocGUI to search and visualise spatial proteomics data. Please refer to the respective documentation and vignettes for full details about the software.
If you use these open-source software for your research, please cite:
Gatto L, Breckels LM, Wieczorek S, Burger T, Lilley KS. Mass-spectrometry-based spatial proteomics data analysis using pRoloc and pRolocdata. Bioinformatics. 2014 May 1;30(9):1322-4. doi:10.1093/bioinformatics/btu013. Epub 2014 Jan 11. PMID:24413670; PMCID:PMC3998135.
Breckels LM, Gatto L, Christoforou A, Groen AJ, Lilley KS, Trotter MW. The effect of organelle discovery upon sub-cellular protein localisation. J Proteomics. 2013 Mar 21. doi:pii: S1874-3919(13)00094-8. 10.1016/j.jprot.2013.02.019. PMID:23523639.
Gatto L., Breckels L.M., Burger T, Nightingale D.J.H., Groen A.J., Campbell C., Mulvey C.M., Christoforou A., Ferro M., Lilley K.S. 'A foundation for reliable spatial proteomics data analysis' Mol Cell Proteomics. 2014 May 20.
Installation
pRolocGUI is written in the Rprogramming language. Before installing the software you need to download R and (optionally) RStudio.
- Download the latest
Rrelease for your operating system from theR website and install it. - Optional, but recommended. Downloadand install the RStudio IDE.
RStudioprovides a good code editor and excellent integration with theRterminal. - Start
RorRStudio. - Install the Bioconductor packages
pRoloc,pRolocdataandpRolocGUI:
pRolocGUI requires R >= 3.1.1 and Bioconductor version >= 3.0. In an R console, type
if (!requireNamespace("BiocManager", quietly=TRUE))
install.packages("BiocManager")
BiocManager::install(c("pRoloc", "pRolocdata", "pRolocGUI"))
Development version
The development code on github can also be installed usingBiocManager::install (or install_github). New pre-release features might not be documented or thoroughly tested and could substantially change prior to release. Use at your own risks.
BiocManager::install("ComputationalProteomicsUnit/pRolocGUI")
Getting started
Before using a package's functionality, it needs to be loaded:
We first load data fromChristoforou et al 2016distributed in the pRolocdata package:
library("pRolocdata")
data(hyperLOPIT2015)
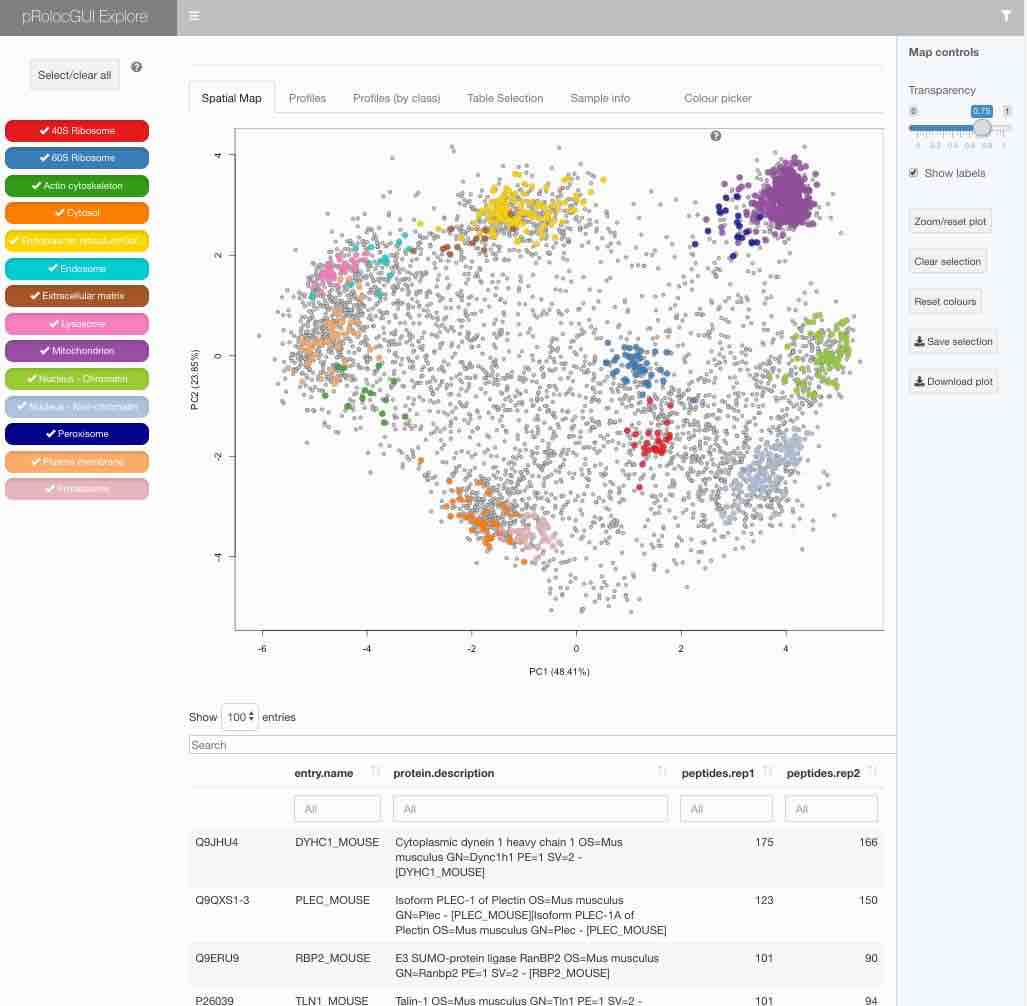
There are 3 different visualisation applications currently available: explore, compare and aggregate. These apps are launched using the pRolocVis function and passing object, which is an MSnSet containing the data one wishes to interrogate. One may also specify which app they wish to use by using the app argument, see ?pRolocVisfor more details. The default app that is loaded ifapp is not specified is the explore application:
pRolocVis(hyperLOPIT2015)
The graphical interface is described in details in the package vignette that is included in the package itself (get it by typingvignette("pRolocGUI") in R), available by clicking the ? once the interface is loaded or can beconsulted online.
More resources
Support
- The Bioconductor support forum
- Open a
pRolocGUIGitHubissue(requires a free GitHub account).
Videos (new videos will appear shortly for the new apps)
- An introduction to Bioconductor
- A brief introduction to pRolocGUI
- Downloading and install R
- Using RStudio
- Installing the pRolocGUI interface
- Starting pRolocGUI - This tutorial is for the older legacy applications. New videos will appear shortly for the new applications.
- Using pRolocGUI to explore and visualise experimental spatial proteomics data - This tutorial is for the older legacy applications. New videos will appear shortly for the new applications.
Tutorial playlist.
General resources
- Teaching material
- R and Bioconductor for proteomicsweb page andpackage.