GitHub - m1frasca/COSNet_GitHub (original) (raw)
COSNet: Cost Sensitive Network for node label prediction on graphs with highly unbalanced labelings
What is COSNet?
COSNet (COst Sensitive neural Network) [1,2] is a novel method to learn node labels biological networks with a high prevalence of negative instances. Examples of this context are the automated prediction of protein functions, the gene disease prioritization and the drug reposition problem.
COSNet is based on a cost-sensitive family of parametric Hopfield networks, whose characteristics can be summarized as follows:
- Class labels and neuron states are conceptually separated. In this way a class of Hopfield networks is introduced, having as parameters the values of neuron states and the neuron thresholds.
- The parameters of the network are learned from the data through an efficient supervised algorithm, in order to take into account the unbalance between positive and negative node labels.
- The dynamics of the network is restricted to its unlabeled part, preserving the minimization of the overall objective function and the a priori information and significantly reducing the time complexity of the learning algorithm.
Architecture
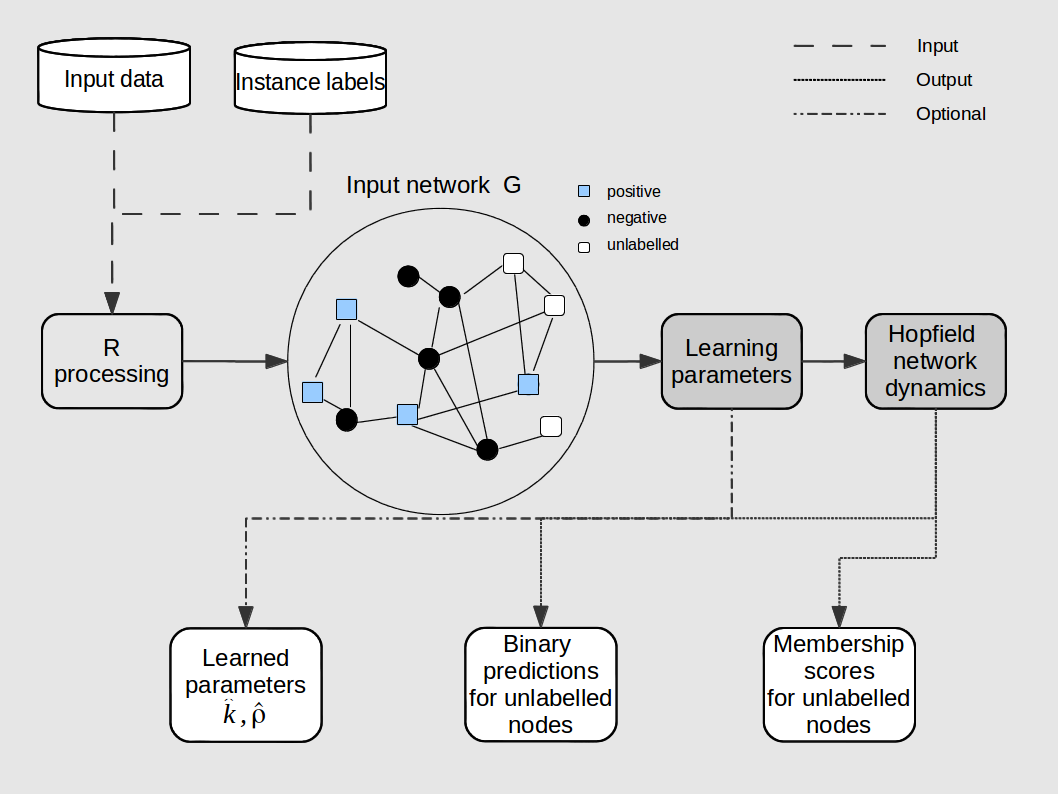
COSNet has three main modules: 1) Data processing. The package provides functions to partition input data in folds (find.division.strat and find.division.not.strat), and to generate temporary labels for the unlabelled instances (generate_labels), to be used in the learning phase. See Section 5.1.1 of reference [1] for details about this step; 2) Learning of parameters. This part of the package realizes the learning procedure of the COSNet algorithm (see Section 5.2 of [1]). The function generate_points projects nodes in the training set into labelled points in the plane (Section 5.2.1), whereas functions optimizep and optimize_pos_above learn the optimal straight line (Section 5.2.2); 3) Network dynamics and regularization. The package provides the function runSubnet to realize the dynamics (with the learned parameters) of the sub-network restricted to unlabelled nodes (Section 5.3 of [1]). Moreover, the function reg_data allows to simulate a regularized dynamics (Section 5.6 of paper [1]).
Installation
COSNet can be installed by running the R environment and typing:
if (!requireNamespace("BiocManager", quietly = TRUE)) install.packages("BiocManager")
BiocManager::install("COSNet")
Try http if https is not available. This will download and install the package. Another possibility is doing it manually. For instance, on a unix/linix R environment, download the package at http://bioconductor.org/packages/release/bioc/src/contrib/COSNet_1.4.1.tar.gz and save it in the current folder. Then from the R prompt type
install.packages("COSNet_1.4.1.tar.gz", repos=NULL)
The COSNet package has no dependencies. Nevertheless, the experiments reported in the package vignette use the R packages bionetdata and PerfMeas, available at the CRAN repository, for loading benchmark data and compute various measure of performance respectively.
The package PerfMeas in turn depends on the Bioconductor R packages limma, graph, and RBGL, which can be installed as described for the COSNet package.
To install the bionetdata package, type
install.packages("bionetdata")
or follow the instruction for the manual installation above.
Example
Here is a simple example. See this link, the manual and the package vignette for more examples.
library(bionetdata);
loading Binary protein-protein interactions from the STRING
data base (von Mering et al. 2002)
data(Yeast.STRING.data)# "Yeast.STRING.data"
FunCat classes annotations (0/1) for the genes included
in Yeast.STRING.data. Annotations refer the funcat-2.1
scheme, and funcat-2.1 data 20070316 data, available from the MIPS web site.
data(Yeast.STRING.FunCat) # "Yeast.STRING.FunCat" labels <- Yeast.STRING.FunCat; labels[labels == 0] <- -1;
excluding the dummy "00" root
labels <- labels[, -which(colnames(labels) == "00")]; n <- nrow(labels); k <- floor(n/10); cat("k = ", k, "\n");
choosing the first class
labeling <- labels[, 1];
randomly choosing a subset of genes whose labels are hidden
hidden <- sort(sample(1:n, k)); hidden.labels <- labeling[hidden]; labeling[hidden] <- 0; out <- COSNet(Yeast.STRING.data, labeling, 0); prediction <- out$pred; TP <- sum(hidden.labels == 1 & prediction == 1); FN <- sum(hidden.labels == 1 & prediction == -1); FP <- sum(hidden.labels == -1 & prediction == 1); out2 <- COSNet(Yeast.STRING.data, labeling, 0.0001); prediction <- out2$pred; TP2 <- sum(hidden.labels == 1 & prediction == 1); FN2 <- sum(hidden.labels == 1 & prediction == -1); FP2 <- sum(hidden.labels == -1 & prediction == 1);
Reference
[1] Frasca, M., Bertoni, A., Re, M. and Valentini, G.
"A neural network algorithm for semi-supervised node label learning from unbalanced data"
Neural Networks, 43, 84-98, 2013.
[2]
Bertoni, A., Frasca, M., Valentini G.
"COSNet: a Cost Sensitive Neural Network for Semi-supervised Learning in Graphs"
In:Machine Learning and Knowledge Discovery in Databases.
European Conference, ECML PKDD 2011, Athens, Greece, Proceedings, Part I,
Lecture Notes in Artificial Intelligence, vol. 6911, pp.219-234, Springer, 2011.