Streamlining Regulon Identification in Bacteria - DOE Joint Genome Institute (original) (raw)
RIViT-seq technology could speed up associating transcription factors with their target genes.
The Science
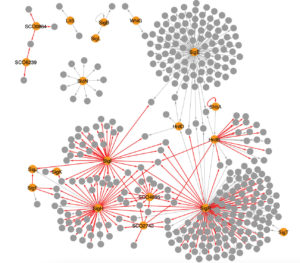
Data yielded from RIViT-seq increased the number of sigma factor-gene pairs confirmed in Streptomyces coelicolor from 209 to 399. Here, grey arrows denote previously known regulation and red arrows are regulation identified by RIViT-seq; orange nodes mark sigma factors while gray nodes mark other genes. (Otani, H., Mouncey, N.J. Nat Commun 13, 3502 (2022). https://doi.org/10.1038/s41467-022-31191-w)
Regulons are a group of genes that can be turned on or off by the same regulatory protein. Researchers demonstrated the efficacy and potential of regulon identification by in vitro transcription-sequencing (RIViT-seq) to facilitate the characterization of transcriptional regulation as well as identification of target genes of transcription factors, particularly sigma factors that initiate transcription in bacteria. Using this technology, which combines an in vitro transcription assay with RNA sequencing, they were able to identify the target genes of 11 sigma factors in Streptomyces coelicolor.
The Impact
As an integrative genome science user facility, connecting genes to functions is part of the mission of the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science User Facility located at Lawrence Berkeley National Laboratory (Berkeley Lab). Transcription factors control when and how genes are turned on or off, so transcriptional regulation is critical to ensuring genes vital for growth and survival across various environments are expressed when their functions are needed.
Data yielded through RIViT-seq expand the known transcriptional regulatory network in bacteria, enabling the discovery of regulatory cascades as well as crosstalk between sigma factors. While characterizing sigma factors is the most straightforward application of the technology, in principle, RIViT-seq can be applied to any transcription factors or regulatory proteins that control transcription.
Summary
Transcription in eukaryotes is handled by three distinct RNA polymerases; in bacteria there is only one type of RNA polymerase responsible for each gene. These bacterial RNA polymerases are made up of a core enzyme complex as well as a sigma factor.
While all bacteria encode at least one principal sigma factor responsible for transcription of housekeeping genes, many bacteria also encode alternative sigma factors to initiate transcription of gene sets associated with functions like stress responses and cellular differentiation.
The tough part is identifying the set of genes associated with any particular factor. Predicting those relationships bioinformatically is almost impossible, and the experimental process for doing so is laborious and complex. RIViT-seq technology simplifies this identification process.
Multiple factors mediate transcription, including activators, repressors and sigma factors. However, currently the process for identifying the particular regulon and target genes associated with a given transcription factor is resource-intensive and unscalable. To test the efficacy of RIViT-seq, researchers recently characterized 12 sigma factors and were able to identify the associated target genes in 11 of those.
That work published in Nature Communications on June 17, 2022, was conducted by JGI Director Nigel Mouncey and research scientist Hiroshi Otani. Both are members of the JGI’s Secondary Metabolite Science Program, which Mouncey leads.
RIViT-seq is a three-step process. The first step is the in vitro transcription assay. The second step consists of RNA sequencing of whole transcriptomes, and the third is determining the 5’-ends by 5’-end sequencing, from which those promoter sequences recognized by a given sigma factor are inferred.
Prior to this most recent study, only 12 sigma factors and 200 genes (or 209 sigma factor-target gene pairs) had been experimentally verified in S. coelicolor. With results from the research, RIViT-seq data have increased the identification of the number of sigma factors linked to at least one target gene to 18, with the number of factor-gene pairs almost doubling to 399. Two of the 11 sigma factors addressed in the study were found to control more than 100 genes, one of which was directly characterized by RIViT-seq.
Further application of RIViT-seq to other transcription factors and even other organisms can fuel a deeper understanding of these transcriptional regulatory networks.
Contacts
BER Contact
Ramana Madupu, Ph.D.
Program Manager
Biological Systems Sciences Division
Office of Biological and Environmental Research
Office of Science
US Department of Energy
[email protected]
PI Contact
Nigel Mouncey
Secondary Metabolite Science Program Lead
Director, DOE Joint Genome Institute
[email protected]
Funding
- This work was supported by the U.S. Department of Energy Office of Science under Contract No. DE-AC02-05CH11231
Publication
- Otani, H., Mouncey, N.J. “RIViT-seq enables systematic identification of regulons of transcriptional machineries.” Nature Communications 13, 3502 (2022). doi. 10.1038/s41467-022-31191-w
Byline: Allison Joy