The mammalian gene function resource: the international knockout mouse consortium (original) (raw)
Abstract
In 2007, the International Knockout Mouse Consortium (IKMC) made the ambitious promise to generate mutations in virtually every protein-coding gene of the mouse genome in a concerted worldwide action. Now, 5 years later, the IKMC members have developed high-throughput gene trapping and, in particular, gene-targeting pipelines and generated more than 17,400 mutant murine embryonic stem (ES) cell clones and more than 1,700 mutant mouse strains, most of them conditional. A common IKMC web portal (www.knockoutmouse.org) has been established, allowing easy access to this unparalleled biological resource. The IKMC materials considerably enhance functional gene annotation of the mammalian genome and will have a major impact on future biomedical research.
Similar content being viewed by others
Introduction
Annotation of the human and mouse genomes has identified more than 20,000 protein-coding genes and more than 3,000 noncoding RNA genes. Together, these genes orchestrate the development and function of the organism from fertilization through embryogenesis to adult life. Despite the dramatic increase in knowledge of variation in human genomes of healthy and diseased individuals, the normal functions of common forms of most genes are still unknown and consequently the disease significance of rare variants remains obscure as well.
To determine gene function, mutation of those genes is required in model organisms. The mouse has long been regarded as ideal for this purpose. Conservation of most aspects of mammalian development, anatomy, metabolism, and physiology between humans and mice is underscored by strong one-to-one orthologous relationships between genes of the two species. Conservation of gene function is strongly supported by similar phenotypic consequences of complete or partial loss-of-function mutations in orthologous genes in both species and by functional replaceability of mouse genes by their human counterparts (Wallace et al. 2007).
To provide a platform for addressing vertebrate gene function on a large scale, the research community came together to establish a genome-wide genetic resource of mouse mutants (Austin et al. 2004; Auwerx et al. 2004). The consensus was that the future currency of this biological resource should be based on ES cells, which can be readily transferred between laboratories and across international boundaries. It was also felt that the most desirable alleles would be those generated by gene targeting. Bespoke designs for each gene would accommodate each gene’s unique structural attributes and take account of adjacent genomic features. Uncertainty in the scalability of gene-targeting technology coupled with the availability of several gene-trap libraries and the speed with which additional mutant alleles could be generated by gene-trapping methods resulted in agreement that the resource should be generated initially by using both gene-targeting and gene-trapping technologies.
Thus, the vision emerged of a core public archive of ES cell clones on a single uniform genetic background, each clone carrying an engineered mutation in a different gene. To extract biological insights from this resource, individual ES cell clones would be converted into mice by individual investigators and organized programs.
To deliver the ES cell resource toward this vision of functional annotation, four international programs in Europe and North America were established with the goal of achieving saturation mutagenesis of the mouse genome: EUCOMM, KOMP, NorCOMM, and TIGM (see Table 3). These programs were the founding members of the International Knockout Mouse Consortium (IKMC), fostering groups to work together in a highly coordinated, standardized manner, to share technologies, to maximize output, and to largely avoid duplication of effort (Collins et al. 2007). The IKMC consortium has generated mainly conditional but also constitutive mutations, with the former class of mutations facilitating tissue-specific assessment of gene function at desired time points, especially in situations where an essential requirement of a gene product in one context can exclude analysis in another.
IKMC technology
The IKMC mutant ES cell resources have been developed for the most part in a C57BL/6N genetic background using cell lines that have achieved clonal germline transmission rates of up to 80 % (Pettitt et al. 2009). Mutations were generated initially by using both gene-trapping and gene-targeting technologies. However, the greater utility and desirability of targeted alleles, designed and generated with nucleotide precision, led to phasing out of gene trapping as the efficiency of the high-throughput gene-targeting pipelines became established. Progressive improvements in mouse genome annotation, computational targeting vector design, and 96-well recombineering protocols as well as high efficiencies of gene targeting have facilitated the rapid construction of targeted ES cell clones at unprecedented rates (Skarnes et al. 2011).
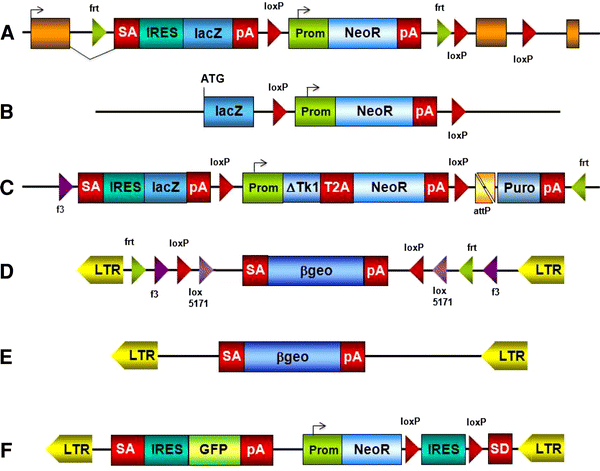
The alleles generated by IKMC members are lacZ tagged and are either null/conditional or null/deletion alleles (Fig. 1). The largest category of targeted clones in the resource contains an allele design known as “knockout-first” from which conditional alleles can be established following exposure to a site-specific recombinase. A conditional allele is created by the deletion of a critical exon which is flanked by loxP sites. Critical exons are those that (1) when deleted, shift the reading frame, (2) are common to all known isoforms, and (3) are contained in the first 50 % of the coding region. Conditional alleles are also amenable to further modification by recombinase-mediated cassette exchange (RMCE), which can be used to insert other coding sequences into these alleles (Osterwalder et al. 2010; Schnütgen et al. 2011). The other major class of mutations in the resource comprises _lacZ_-tagged nulls, constructed as large deletions that are not amenable to further modification (Valenzuela et al. 2003).
Fig. 1
Vectors used by the IKMC: targeting vectors: a EUCOMM/KOMP-CSD knockout-first allele; b KOMP-Regeneron null allele generating large deletions; c NorCOMM promoter-driven targeting vector. Most commonly used trapping vectors: d conditional EUCOMM vector rsFlipROSAβgeo*; e TIGM vector VICTR76; f NorCOMM vector UPA
IKMC ES cell and mouse resources
Currently, the IKMC ES cell resource contains targeted and trapped alleles for 17,473 unique protein-coding genes. The targeted ES cell resource contains mutations in 13,840 unique genes of which 10,100 are null/conditional. The ES cell gene-trap resource contains mutations in 11,539 unique genes of which 4,414 are conditional (Table 1, www.knockoutmouse.org). Due to the random nature of gene trapping, there is some overlap and redundancy between the resources. In general, the IKMC has generated a median of four independent clones per gene. This redundancy not only helps to assure germline transmission but also provides significant allelic diversity (e.g., conditional or nonconditional alleles, different insertion sites, and vector designs). The use of 129 Sv ES cells early in the project for the gene-trap resource also provides the option of studying mutations in the same gene in two different genetic backgrounds for 4,600 genes in the resource.
Table 1 IKMC progress (as of February 2012)
The process of converting IKMC ES cell resources into mice was initiated alongside the gene-targeting and -trapping pipelines as a systematic quality control measure and also for phenotyping purposes. So far, 1,709 mutant mouse lines have been established by all the IKMC members (Table 1). Generation of mice at scale in addition has taken place in organized efforts in a few centers, coupled with systematic phenotyping programs such as the EC-supported EUMODIC Project and the Wellcome Trust Sanger Institute’s Mouse Genetics Programme (MGP) (Table 3). As of July 2012, from both projects’ databases, i.e., www.europhenome.org and www.sanger.ac.uk/mouseportal, already 919 targeted lines can be identified that have been phenotypically examined. Of these, 652 have an annotated phenotype.
IKMC repositories and distribution
Repositories have an essential function to secure and maintain the investment in the resource for future generations of scientists. Repositories have the obligation to ensure and preserve the quality of the resource and to act as honest brokers with efficient, unrestricted, and unencumbered distribution to third-party users.
The distribution of IKMC vectors and ES cells takes place through several repositories in Europe, the US, and Canada (EuMMCR, KOMP, TIGM, and CMMR, Tables 2, 3). The repositories conduct stringent quality control on the vectors and ES cell clones prior to distribution to ensure the integrity of each allele. They currently distribute nonoverlapping sectors of the IKMC resource to a global user group. Altogether, these centers have already distributed 1,029 targeting vectors and 4,126 mutant ES cell alleles.
Table 2 IKMC reagents distributed (as of February 2012)
Table 3 Relevant IKMC web sites
The IKMC mice are distributed through EMMA, MMRRC/KOMP, TIGM, and CMMR, the European, US, and Canadian mouse repositories, respectively (Tables 2, 3). To date, of the 1,709 mutated mouse lines, 1,107 (65 %) have been distributed outside of the center in which they were initially produced (Table 2). The dominant impact of IKMC resources on the activity of these repositories is already apparent, e.g., more than 50 % of mice distributed by EMMA were derived from IKMC resources. IKMC mice are also being generated and analyzed in distributed activities by hundreds of individual specialist laboratories worldwide, which to date have cumulatively received 4,126 mutant ES cell alleles from the repositories.
In summary, to date already 6,262 mutant alleles have been distributed worldwide as vectors, ES cells, or mice (about 35 % of IKMC alleles available), and the international requests are still increasing.
IKMC web portal
The integrated, public IKMC web portal (http://www.knockoutmouse.org) summarizes the IKMC progress and enables researchers to obtain IKMC genetic resources from designated repositories. It links to IKMC members’ web pages and to related genetic resources such as those available from the International Gene Trap Consortium (IGTC) and the International Mouse Strain Resource (IMSR) (Table 3) (Ringwald et al. 2011). The IKMC alleles are displayed on the Ensembl and UCSC genome browsers with standardized allele identification registered in Mouse Genome Informatics (MGI, www.informatics.jax.org) database.
The IKMC web portal supports use of the genetic resources through provision of detailed molecular structures of mutant alleles. Researchers may nominate genes of interest for prioritization if targeted mutations are not yet available, although the current coverage of 87 % of the protein-coding genes provides a high chance that a useful allele has already been generated.
IKMC and beyond
The IKMC ES cell resource was envisaged as an essential stepping stone required for the efficient generation of mouse mutants at scale. This resource is nearing completion and is already being accessed by novel large-scale programs to generate mice for phenotypic analysis. An organized effort, the International Mouse Phenotyping Consortium (IMPC), has just been launched (Table 3). Many centers worldwide will contribute to this effort, ultimately generating and analyzing more than 15,000 mouse mutants based on the IKMC resource over the next decade, establishing what will constitute a functional Encyclopedia of the Mammalian Genome.
The IKMC resource, although very comprehensive, still remains incomplete. Ongoing efforts, performed mainly by the EUCOMMTools project (Table 3), will improve genome coverage and the quality of the mutant alleles in the resource over the next 3 years. This will include replacing gene-trap alleles with targeted ones and generating conditional alleles when only a null allele is available. The resource will also continue to expand to cover other classes of genes such as noncoding RNAs.
The IKMC resource includes both vectors and ES cells that can be further modified by RMCE to insert different coding sequences into any conditional mutant IKMC allele such as reporter genes, recombinases, and human orthologs or, e.g., an allele’s missense mutations. In an era in which an extraordinary amount of human variation sequence is becoming available, functional assessment of variants will require analysis under physiological levels of expression and regulation, most reliably achieved by insertion into the mouse orthologous locus.
The examination of a null allele is the requisite first step in order to establish the ground state (null phenotype) of any gene function in the genome. Subsequently, it is desirable to conduct more focused analysis by controlling the generation of the mutation temporally and spatially. The conditional sector of the resource is designed for use with the recombinase Cre, for which a number of transgenic mouse lines are available. The number and characterization level of these lines will increase with various initiatives currently underway [like EUCOMMTools, CanEuCre, and the NorCOMM successor project NorCOMM2LS (Table 3)], and will eventually cover the majority of embryonic and adult tissues and cell types.
Phenotypic analysis of mutant mouse lines generated from IKMC resources enables a better understanding of complex biology such as whole organism physiology, behavior, and adult tissue integrity. In addition, biological insights can also be gained from analysis of cellular phenotypes in culture. IKMC resources facilitate the generation of homozygous mutant ES cells by retargeting the second allele in vitro.
Genetic manipulation of mouse ES cells has reached such a degree of efficiency and sophistication that ES cells can be generated with virtually any genetic change. The success of these technologies provides the groundwork for extension to other mammalian species such as rat, and to human ES and induced pluripotent stem (iPS) cells.
References
- Austin CP, Battey JF, Bradley A, Bucan M, Capecchi M, Collins FS, Dove WF, Duyk G, Dymecki S, Eppig JT et al (2004) The knockout mouse project. Nat Genet 36:921–924
Article PubMed CAS Google Scholar - Auwerx J, Avner P, Baldock R, Ballabio A, Balling R, Barbacid M, Berns A, Bradley A, Brown S, Carmeliet P et al (2004) The European dimension for the mouse genome mutagenesis program. Nat Genet 36:925–927
Article PubMed CAS Google Scholar - Collins FS, Rossant J, Wurst W (2007) A mouse for all reasons. Cell 128:9–13
Article PubMed CAS Google Scholar - Osterwalder M, Galli A, Rosen B, Skarnes WC, Zeller R, Lopez-Rios J (2010) Dual RMCE for efficient re-engineering of mouse mutant alleles. Nat Methods 7:893–895
Article PubMed CAS Google Scholar - Pettitt SJ, Liang Q, Rairdan XY, Moran JL, Prosser HM, Beier DR, Lloyd KC, Bradley A, Skarnes WC (2009) Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat Methods 6:493–495
Article PubMed CAS Google Scholar - Ringwald M, Iyer V, Mason JC, Stone KR, Tadepally HD, Kadin JA, Bult CJ, Eppig JT, Oakley DJ, Briois S et al (2011) The IKMC web portal: a central point of entry to data and resources from the International Knockout Mouse Consortium. Nucleic Acids Res 39:D849–D855
Article PubMed Google Scholar - Schnütgen F, Ehrmann F, Poser I, Hubner NC, Hansen J, Floss T, deVries I, Wurst W, Hyman A, Mann M, von Melchner H (2011) Resources for proteomics in mouse embryonic stem cells. Nat Methods 8:103–104
Article PubMed Google Scholar - Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T et al (2011) A conditional knockout resource for genome-wide analysis of mouse gene function. Nature 474:337–342
Article PubMed CAS Google Scholar - Valenzuela DM, Murphy AJ, Frendewey D, Gale NW, Econonomides AN, Auerbach W, Poueymirou WT, Adams NC, Rojas J, Yasenchak J et al (2003) High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat Biotechnol 21:652–659
Article PubMed CAS Google Scholar - Wallace HAC, Marques-Kranc F, Richardson M, Luna-Crespo F, Sharpe JA, Hughes J, Wood WG, Higgs DR, Smith AJH (2007) Manipulating the mouse genome to engineer precise functional syntenic replacements with human sequence. Cell 128:197–209
Article PubMed CAS Google Scholar
Acknowledgments
The authors thank the European Commission (EC, LSHG-CT-2005-018931, Health-F4-2010-261492), the National Institutes of Health (NIH), Genome Canada, and the State of Texas for funding.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
- The Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, CB10 1HH, UK
Allan Bradley, Joanna Bottomley, Wendy Bushell, Richard Houghton, Vivek Iyer, Karen Kennedy, Manousos Koutsourakis, Alejandro O. Mujica, Ramiro Ramirez-Solis, Barry Rosen, Ed Ryder, William C. Skarnes, Damian Smedley & Anthony P. West - Biotechnology Center (BIOTEC) of the Technische Universität Dresden, 01307, Dresden, Germany
Konstantinos Anastassiadis & A. Francis Stewart - Institut Clinique de la Souris and Institut de Génétique et de Biologie Moléculaire et Cellulaire, 67404, Illkirch Cedex, France
Abdelkader Ayadi, Marie-Christine Birling, Yann Hérault, Guillaume Pavlovic & Mohammed Selloum - National Institute on Deafness and Other Communication Disorders (NIH), Bethesda, MD, 20892, USA
James F. Battey - Genome Canada, Ottawa, ON, K2P 1P1, Canada
Cindy Bell & Kate Swan - Mammalian Genetics Unit, MRC Harwell, Harwell Science and Innovation Campus, Oxfordshire, OX11 0RD, UK
Steve D. Brown, Martin Fray & Lydia Teboul - Institute of Developmental Genetics, Helmholtz Zentrum München, Technische Universität München, 85764, Neuherberg, Germany
Antje Bürger, Roland H. Friedel, Jens Hansen, Andreas Hörlein, Cornelia Kaloff, Joel Zupicich Schick, Claudia Seisenberger & Wolfgang Wurst - The Jackson Laboratory, Bar Harbor, ME, 04609, USA
Carol J. Bult, Janan T. Eppig, James A. Kadin, Jeremy Mason, Martin Ringwald, Kevin Stone & Hamsa Tadepally - National Institutes of Health, Bethesda, MD, 20892, USA
Francis S. Collins - Infectious Diseases and Public Health, European Commission, DG Research & Innovation, 1049, Brussels, Belgium
Christian Desaintes & Jacques E. Remacle - Istituto di Biologia Cellulare, Consiglio Nazionale delle Ricerche (CNR), Monterotondo-Scalo, 00015, Rome, Italy
Brendan Doe, Marcello Raspa & Glauco P. Tocchini-Valentini - Velocigene Division, Regeneron Pharmaceuticals Inc., Tarrytown, NY, 10591, USA
Aris Economides, David Frendewey, Alejandro O. Mujica & David Valenzuela - The Texas A&M Institute for Genomic Medicine, College Station, TX, 77843-4485, USA
Richard H. Finnell & Michael P. McLeod - University of Texas at Austin, Austin, TX, 78712, USA
Richard H. Finnell - National Institutes of Health, Bethesda, MD, 20205, USA
Colin Fletcher, Mark Moore & Jane L. Peterson - Department of Cell Biology, Center of Biomedical Genetics, Erasmus University Medical Center, 3015 GE, Rotterdam, The Netherlands
Frank G. Grosveld - Manitoba Institute of Cell Biology, University of Manitoba, Winnipeg, MB, R3E OV9, Canada
Geoffrey Hicks - Institute of Experimental Genetics, Helmholtz Zentrum München, 85764, Neuherberg, Germany
Martin Hrabé de Angelis & Susan Marschall - Department of Development and Regeneration, Faculty of Medicine, University of Leuven (KU Leuven), 3000, Leuven, Belgium
Danny Huylebroeck - Children’s Hospital Oakland Research Institute (CHORI), Oakland, CA, 94609, USA
Pieter J. de Jong, Mikhail Nefedov & Yuko Yoshinaga - Mouse Biology Program, School of Veterinary Medicine, University of California, Davis, CA, 95616, USA
K. C. Kent Lloyd - Research Institute, The Hospital for Sick Children, SickKids Foundation, Toronto, ON, M5G2L3, Canada
Colin McKerlie, Lauryl M. Nutter & Janet Rossant - Department of Molecular Haematology, University of Frankfurt Medical School, 60590, Frankfurt am Main, Germany
Harald von Melchner & Frank Schnütgen - Samuel Lunenfeld Research Institute, Mount Sinai Hospital, Joseph and Wolf Lebovic Health Complex, Toronto, ON, M5G 1X5, Canada
Andras Nagy - Division of Basic Neuroscience and Research, National Institute of Drug Abuse (NIDA), Bethesda, MD, 20892-0001, USA
Jonathan Pollock - Department of Biochemistry and Molecular Biology, University of Calgary, Calgary, AB, T2N 1N4, Canada
Derrick E. Rancourt - European Molecular Biology Laboratory (EMBL), Monterotondo, 00015, Rome, Italy
Nadia Rosenthal - Centre for Cardiovascular Research, Department of Vertebrate Genomics, Charité, 10115, Berlin, Germany
Patricia Ruiz Noppinger - Department of Physiology, Development and Neuroscience, University of Cambridge, Cambridge, CB2 3EG, UK
Paul Schofield - Department of Medical Genetics, Centre for Molecular Medicine and Therapeutics at the Child & Family Research Institute, University of British Columbia, Vancouver, BC, V5Z 4H4, Canada
Elizabeth M. Simpson - European Bioinformatics Institute (EBI), Hinxton, Cambridge, CB10 1ST, UK
Damian Smedley - The Ottawa Hospital Research Institute, Ottawa, ON, K1N 6N5, Canada
William L. Stanford - Division of Developmental Genetics, Center for Animal Resources and Development, Institute of Resource Development and Analysis, Kumamoto University, Kumamoto, 860-0811, Japan
Ken-ichi Yamamura - Icahn Medical Institute, The Mount Sinai Hospital, New York, NY, 10029, USA
Roland H. Friedel - Max-Planck-Institute of Psychiatry, 80804, Munich, Germany
Wolfgang Wurst - Deutsches Zentrum fuer Neurodegenerative Erkrankungen e.V. (DZNE) Site Munich, 80336, Munich, Germany
Wolfgang Wurst
Authors
- Allan Bradley
You can also search for this author inPubMed Google Scholar - Konstantinos Anastassiadis
You can also search for this author inPubMed Google Scholar - Abdelkader Ayadi
You can also search for this author inPubMed Google Scholar - James F. Battey
You can also search for this author inPubMed Google Scholar - Cindy Bell
You can also search for this author inPubMed Google Scholar - Marie-Christine Birling
You can also search for this author inPubMed Google Scholar - Joanna Bottomley
You can also search for this author inPubMed Google Scholar - Steve D. Brown
You can also search for this author inPubMed Google Scholar - Antje Bürger
You can also search for this author inPubMed Google Scholar - Carol J. Bult
You can also search for this author inPubMed Google Scholar - Wendy Bushell
You can also search for this author inPubMed Google Scholar - Francis S. Collins
You can also search for this author inPubMed Google Scholar - Christian Desaintes
You can also search for this author inPubMed Google Scholar - Brendan Doe
You can also search for this author inPubMed Google Scholar - Aris Economides
You can also search for this author inPubMed Google Scholar - Janan T. Eppig
You can also search for this author inPubMed Google Scholar - Richard H. Finnell
You can also search for this author inPubMed Google Scholar - Colin Fletcher
You can also search for this author inPubMed Google Scholar - Martin Fray
You can also search for this author inPubMed Google Scholar - David Frendewey
You can also search for this author inPubMed Google Scholar - Roland H. Friedel
You can also search for this author inPubMed Google Scholar - Frank G. Grosveld
You can also search for this author inPubMed Google Scholar - Jens Hansen
You can also search for this author inPubMed Google Scholar - Yann Hérault
You can also search for this author inPubMed Google Scholar - Geoffrey Hicks
You can also search for this author inPubMed Google Scholar - Andreas Hörlein
You can also search for this author inPubMed Google Scholar - Richard Houghton
You can also search for this author inPubMed Google Scholar - Martin Hrabé de Angelis
You can also search for this author inPubMed Google Scholar - Danny Huylebroeck
You can also search for this author inPubMed Google Scholar - Vivek Iyer
You can also search for this author inPubMed Google Scholar - Pieter J. de Jong
You can also search for this author inPubMed Google Scholar - James A. Kadin
You can also search for this author inPubMed Google Scholar - Cornelia Kaloff
You can also search for this author inPubMed Google Scholar - Karen Kennedy
You can also search for this author inPubMed Google Scholar - Manousos Koutsourakis
You can also search for this author inPubMed Google Scholar - K. C. Kent Lloyd
You can also search for this author inPubMed Google Scholar - Susan Marschall
You can also search for this author inPubMed Google Scholar - Jeremy Mason
You can also search for this author inPubMed Google Scholar - Colin McKerlie
You can also search for this author inPubMed Google Scholar - Michael P. McLeod
You can also search for this author inPubMed Google Scholar - Harald von Melchner
You can also search for this author inPubMed Google Scholar - Mark Moore
You can also search for this author inPubMed Google Scholar - Alejandro O. Mujica
You can also search for this author inPubMed Google Scholar - Andras Nagy
You can also search for this author inPubMed Google Scholar - Mikhail Nefedov
You can also search for this author inPubMed Google Scholar - Lauryl M. Nutter
You can also search for this author inPubMed Google Scholar - Guillaume Pavlovic
You can also search for this author inPubMed Google Scholar - Jane L. Peterson
You can also search for this author inPubMed Google Scholar - Jonathan Pollock
You can also search for this author inPubMed Google Scholar - Ramiro Ramirez-Solis
You can also search for this author inPubMed Google Scholar - Derrick E. Rancourt
You can also search for this author inPubMed Google Scholar - Marcello Raspa
You can also search for this author inPubMed Google Scholar - Jacques E. Remacle
You can also search for this author inPubMed Google Scholar - Martin Ringwald
You can also search for this author inPubMed Google Scholar - Barry Rosen
You can also search for this author inPubMed Google Scholar - Nadia Rosenthal
You can also search for this author inPubMed Google Scholar - Janet Rossant
You can also search for this author inPubMed Google Scholar - Patricia Ruiz Noppinger
You can also search for this author inPubMed Google Scholar - Ed Ryder
You can also search for this author inPubMed Google Scholar - Joel Zupicich Schick
You can also search for this author inPubMed Google Scholar - Frank Schnütgen
You can also search for this author inPubMed Google Scholar - Paul Schofield
You can also search for this author inPubMed Google Scholar - Claudia Seisenberger
You can also search for this author inPubMed Google Scholar - Mohammed Selloum
You can also search for this author inPubMed Google Scholar - Elizabeth M. Simpson
You can also search for this author inPubMed Google Scholar - William C. Skarnes
You can also search for this author inPubMed Google Scholar - Damian Smedley
You can also search for this author inPubMed Google Scholar - William L. Stanford
You can also search for this author inPubMed Google Scholar - A. Francis Stewart
You can also search for this author inPubMed Google Scholar - Kevin Stone
You can also search for this author inPubMed Google Scholar - Kate Swan
You can also search for this author inPubMed Google Scholar - Hamsa Tadepally
You can also search for this author inPubMed Google Scholar - Lydia Teboul
You can also search for this author inPubMed Google Scholar - Glauco P. Tocchini-Valentini
You can also search for this author inPubMed Google Scholar - David Valenzuela
You can also search for this author inPubMed Google Scholar - Anthony P. West
You can also search for this author inPubMed Google Scholar - Ken-ichi Yamamura
You can also search for this author inPubMed Google Scholar - Yuko Yoshinaga
You can also search for this author inPubMed Google Scholar - Wolfgang Wurst
You can also search for this author inPubMed Google Scholar
Corresponding authors
Correspondence toAllan Bradley or Wolfgang Wurst.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Bradley, A., Anastassiadis, K., Ayadi, A. et al. The mammalian gene function resource: the international knockout mouse consortium.Mamm Genome 23, 580–586 (2012). https://doi.org/10.1007/s00335-012-9422-2
- Received: 06 April 2012
- Accepted: 20 July 2012
- Published: 12 September 2012
- Issue Date: October 2012
- DOI: https://doi.org/10.1007/s00335-012-9422-2