Bridging Islands of Information to Establish an Integrated Knowledge Base of Drugs and Health Outcomes of Interest (original) (raw)
Abstract
The entire drug safety enterprise has a need to search, retrieve, evaluate, and synthesize scientific evidence more efficiently. This discovery and synthesis process would be greatly accelerated through access to a common framework that brings all relevant information sources together within a standardized structure. This presents an opportunity to establish an open-source community effort to develop a global knowledge base, one that brings together and standardizes all available information for all drugs and all health outcomes of interest (HOIs) from all electronic sources pertinent to drug safety. To make this vision a reality, we have established a workgroup within the Observational Health Data Sciences and Informatics (OHDSI, http://ohdsi.org) collaborative. The workgroup’s mission is to develop an open-source standardized knowledge base for the effects of medical products and an efficient procedure for maintaining and expanding it. The knowledge base will make it simpler for practitioners to access, retrieve, and synthesize evidence so that they can reach a rigorous and accurate assessment of causal relationships between a given drug and HOI. Development of the knowledge base will proceed with the measureable goal of supporting an efficient and thorough evidence-based assessment of the effects of 1,000 active ingredients across 100 HOIs. This non-trivial task will result in a high-quality and generally applicable drug safety knowledge base. It will also yield a reference standard of drug–HOI pairs that will enable more advanced methodological research that empirically evaluates the performance of drug safety analysis methods.
FormalPara Key Points
| The individuals who possess the expertise to synthesize evidence on a medication’s safety are hindered by numerous disconnected “islands of information” |
|---|
| A workgroup within the Observational Health Data Sciences and Informatics (OHDSI, http://ohdsi.org) collaborative is addressing this issue by establishing an open-source community effort to develop a global knowledge base that brings together and standardizes all available information for all drugs and all health outcomes of interest from all electronic sources pertinent to drug safety |
| Striving toward the goal of a generally useful knowledge base, though ambitious, is necessary for advancing the science of drug safety because it will make it simpler for practitioners to access, retrieve, and synthesize evidence so that they can reach a rigorous and accurate assessment of causal relationships between a given drug and the health outcome of interest |
1 Introduction
“The investigator is staggered by the findings and conclusions of thousands of other workers—conclusions which he cannot find time to grasp, much less to remember, as they appear.”—Bush 1945 [1]
When Dr. Vannevar Bush penned this lament 7 decades ago, the then Director of the United States Office of Scientific Research and Development was calling post-World War II scientists to conduct research that would yield a revolutionary approach to representing and retrieving information. At the time, distributed document collections and taxonomic indexing schemes were hindering the ability of researchers to identify important connections that could yield new scientific insights. The Internet, electronic document collections, hypertext, advanced information retrieval systems, and digital social networks are some of the many advances since Dr. Bush first articulated his vision. Unfortunately, his lament still resonates with the contemporary drug safety practitioner. Today, an overwhelming amount of drug safety-relevant information is being generated and stored in a wide array of disparate information sources using differing terminologies at a faster pace than ever before. Product manufacturers, regulatory agencies, and prescribers have an obligation to the public to correctly interpret and properly act on this information in a timely manner. However, the individuals who possess the expertise to synthesize evidence on a medication’s safety are hindered by numerous disconnected “islands of information.”
Like a photo mosaic, a clear and understandable image of a potential drug safety issue can emerge when the relevant sources of evidence are brought together. The written protocol for a pre-marketing drug trial can help determine if an adverse event mentioned in a spontaneous report is causally related to the drug exposure or the condition being treated. A well-designed observational study using electronic health records data can suggest what categories of patients would be most at risk for developing an adverse drug reaction listed in product labeling. A published case report can add credence to a potential drug–adverse event association identified by mining spontaneous reporting data or longitudinal observational health databases. A systematic review of clinical trials testing a drug’s efficacy for an off-label indication can provide data on adverse events that can occur in populations not mentioned in drug product labeling. A knowledge base (KB) of drug pharmacological properties and molecular targets can yield information useful for inferring the biological plausibility of a suspected drug-related adverse event.
Unfortunately, the information from these and many other potentially useful sources is stored in different systems with distinct information formats, employing non-interoperable terminology schemes, and requiring unique skills to navigate and explore (Table 1). This situation makes it extremely time consuming and resource intensive to retrieve the necessary information when conducting a comprehensive assessment of a potential safety signal. The investigation of drug safety concerns tends to be manual, highly iterative, with a steep learning curve, and perpetually at risk for errors of omission due to the complexities involved in searching across multiple domains for related information.
Table 1 A sample of sources of information that could potentially contain evidence relevant to a suspected association between a drug and a health outcome of interest
The entire drug safety enterprise has a need to search, retrieve, evaluate, and synthesize scientific evidence more efficiently. This presents a tremendous opportunity to establish an open-source community effort to develop a global KB, one that brings together and standardizes all available information for all drugs and all health outcomes of interest (HOIs) from all electronic sources pertinent to drug safety. The community needs to go beyond simply enabling cross-resource queries to establish an empirical evidence base about the reliability of information sources used in the drug safety assessment process.
The quote by Dr. Vannevar Bush at the beginning of this paper is taken from a paper in which he invited post-war scientists to use emerging technologies such as photocells, cathode ray tubes, and “arithmetical machines” (very early computers) to make the ever growing scientific record much more natural to synthesize. Were he alive today, he might suggest relatively recent technologies such as biomedical ontologies [[2](/article/10.1007/s40264-014-0189-0#ref-CR2 "Yu AC. Methods in biomedical ontology. J Biomed Inform. 2006;39(3):252–66. doi: 10.1016/j.jbi.2005.11.006
.")\], Semantic Web Linked Data \[[3](/article/10.1007/s40264-014-0189-0#ref-CR3 "Marshall MS, Boyce R, Deus HF, Zhao J, Willighagen EL, Samwald M, et al. Emerging practices for mapping and linking life sciences data using RDF—a case series. Web Semant Sci Serv Agents World Wide Web. 2012;14:2–13.")\], natural language processing, and machine learning. Biomedical ontologies and Semantic Web Linked Data would be recommended for their potential to enable all sources to be integrated in a way that allows for both summative queries (e.g., “How many data sources suggest that drug X is associated with HOI Y?”) and the ability to “drill down” into specific data sources (e.g., “When did source A first suggest that drug X is associated with HOI Y?”); natural language processing would be recommended for its potential to enable the addition of knowledge mentioned within the text documents (e.g., adverse drug reactions recorded in tables and sections of drug product labeling); and machine learning would be recommended for its potential to automate much of the process for identifying positive and negative drug–HOI associations. Moreover, innovative sources of drug safety evidence, such as inferences derived from predictive methods emerging from the nascent field of network medicine \[[4](/article/10.1007/s40264-014-0189-0#ref-CR4 "Jacunski A, Tatonetti NP. Connecting the dots: applications of network medicine in pharmacology and disease. Clin Pharmacol Therap. 2013;94(6):659–69. doi:
10.1038/clpt.2013.168
.")\] and weblogs \[[5](/article/10.1007/s40264-014-0189-0#ref-CR5 "Yeleswarapu S, Rao A, Joseph T, Saipradeep VG, Srinivasan R. A pipeline to extract drug-adverse event pairs from multiple data sources. BMC Med Inform Decis Mak. 2014;14(1):13.")\], should be considered as potentially valuable additional forms of evidence.To make this vision a reality, we have established a workgroup within the Observational Health Data Sciences and Informatics (OHDSI, http://ohdsi.org) collaborative. The workgroup’s mission is to establish an open-source standardized KB for the effects of medical products and an efficient semi-automated procedure for maintaining and expanding it.
2 A Focal Point for the Integration of Information Sources Relevant to Drug Safety
We believe that development of the proposed KB should proceed with the measureable goal of supporting an efficient and thorough evidence-based assessment of the effects of 1,000 active ingredients across 100 HOIs. This non-trivial task will result in a high-quality and generally applicable drug safety KB, providing a focal point to guide design decisions. These include what information sources to include, what terminologies to employ, how to handle data that comes with uncertainty (e.g., associations mined from spontaneous reports, risks identified in pharmacoepidemiological studies, or the output of processing the scientific literature using natural language processing algorithms), and how to accommodate conflicting evidence. The large-scale evidence assessment task will also be a major contribution to the global drug safety research community because it will yield a reference standard of drug–HOI pairs that will enable more advanced methodological research that empirically evaluates the performance of drug safety analysis methods.
The target of 1,000 drugs is motivated by the fact that this number represents a significant proportion of the drugs used in practice. At the time of this writing, we estimate that it would represent 64 % of the 1,565 unique active ingredients listed in the drugs@FDA database as currently marketed for prescription or over-the-counter use in the USA (though the choice of drugs will not be limited to a single country’s market). The choice of 100 HOIs is motivated by the fact that the number is sufficiently greater than previous efforts so as to spur innovative approaches to making the drug–HOI assessments more efficient. The specific list of drugs and HOIs will include those already examined in previous references standards and those considered to be high priority by our pharmacovigilance collaborators. We will further extend the drug list to ensure a representative sample, taking into account such attributes as marketing duration, pharmacological class, and prevalence of exposure. Similarly, we will choose additional HOIs so as to ensure an accurate representation of severity, system/organ class, and likelihood of mention in various sources.
2.1 The Broad Utility of a Drug Safety KB
Considering a given HOI, one of a drug safety practitioner’s main tasks is to search for all relevant evidence for a positive or negative association between any drug and the HOI and synthesize that evidence to make a final judgment on the veracity of the association. Practitioners routinely need to review disparate information from scientific literature, product labeling, spontaneous adverse reports, observational health data, and other sources. This discovery and synthesis process would be greatly accelerated through access to a common framework that brings all of these information sources together within a standardized structure.
It is also quite possible that the KB will have value beyond drug safety; product manufacturers may use the information to assess areas of unmet medical need or identify targets for drug re-purposing, providers may use this information to support clinical decisions, and patients may benefit from access to a standard, easy-to-use interface that provides consistent information about their treatments and their potential effects. Moreover the OHDSI KB will directly impact methodological research and empirical evaluation of drug safety methods by enabling the development of a globally acceptable drug–HOI reference set.
2.2 The Need for a Globally Acceptable Drug–HOI Reference Standard
Over the past decade, a number of experiments have been performed to estimate the ability of drug safety analysis methods to discriminate between drugs causally related with specific HOIs (drug–HOI “positive controls”) and drugs that have no causal relation (drug–HOI “negative controls”), measure the expected time to detection, and quantify the magnitude of error that should be anticipated from any effect estimate [[6](/article/10.1007/s40264-014-0189-0#ref-CR6 "DuMouchel W, Ryan PB, Schuemie MJ, Madigan D. Evaluation of disproportionality safety signaling applied to healthcare databases. Drug Saf. 2013;36(Suppl 1):S123–32. doi: 10.1007/s40264-013-0106-y
.")–[20](/article/10.1007/s40264-014-0189-0#ref-CR20 "Norén GN, Bergvall T, Ryan PB, Juhlin K, Schuemie MJ, Madigan D. Empirical performance of the calibrated self-controlled cohort analysis within temporal pattern discovery: lessons for developing a risk identification and analysis system. Drug Saf. 2013;36(Suppl 1):S107–21. doi:
10.1007/s40264-013-0095-x
.")\]. The primary means for conducting these methodological experiments is to perform a retrospective evaluation that compares the results from the drug safety analysis process with some pre-defined reference standard. Ideally, a reference standard would represent a large collection of drug–HOI combinations, be based on complete and certain information about the strength of association, and provide the provenance (e.g., source and date of creation) of evidence items used to develop the standard. In practice, the task of establishing a reference standard involves resource-intensive information gathering and decision-making under uncertainty.To illustrate the varying approaches to creating a reference set, Table 2 highlights the evidence sources and sampling frame from five recent methodological experiments where drug–HOI reference sets were developed. The reference standards developed by Hochberg et al. [[19](/article/10.1007/s40264-014-0189-0#ref-CR19 "Hochberg AM, Hauben M, Pearson RK, O’Hara DJ, Reisinger SJ, Goldsmith DI, et al. An evaluation of three signal-detection algorithms using a highly inclusive reference event database. Drug Saf. 2009;32(6):509–25. doi: 10.2165/00002018-200932060-00007
.")\] and Alvarez et al. \[[17](/article/10.1007/s40264-014-0189-0#ref-CR17 "Alvarez Y, Hidalgo A, Maignen F, Slattery J. Validation of statistical signal detection procedures in eudravigilance post-authorization data: a retrospective evaluation of the potential for earlier signalling. Drug Saf. 2010;33(6):475–87. doi:
10.2165/11534410-000000000-00000
.")\] were initially used to support evaluation of spontaneous adverse event reporting analyses, whereas the Observational Medical Outcomes Partnership (OMOP) \[[8](/article/10.1007/s40264-014-0189-0#ref-CR8 "Ryan PB, Madigan D, Stang PE, Overhage JM, Racoosin JA, Hartzema AG. Empirical assessment of methods for risk identification in healthcare data: results from the experiments of the Observational Medical Outcomes Partnership. Stat Med. 2012;31(30):4401–15. doi:
10.1002/sim.5620
."), [21](/article/10.1007/s40264-014-0189-0#ref-CR21 "Ryan PB, Schuemie MJ, Welebob E, Duke J, Valentine S, Hartzema AG. Defining a reference set to support methodological research in drug safety. Drug Saf. 2013;36(Suppl 1):S33–47. doi:
10.1007/s40264-013-0097-8
.")\] and Exploring and Understanding Adverse Drug Reactions by Integrative Mining of Clinical Records and Biomedical Knowledge (EU-ADR) \[[22](/article/10.1007/s40264-014-0189-0#ref-CR22 "Coloma PM, Avillach P, Salvo F, Schuemie MJ, Ferrajolo C, Pariente A, et al. A reference standard for evaluation of methods for drug safety signal detection using electronic healthcare record databases. Drug Saf. 2013;36(1):13–23. doi:
10.1007/s40264-012-0002-x
.")\] reference sets were designed to facilitate research in observational health databases. What is most striking in this summary is that the different approaches employed to select and evaluate drug–HOI cases resulted in heterogeneous reference standards with different degrees of confidence in the final output.Table 2 Reference sets established to support methodological research in drug safety
Table 3 Hypothetical output of the knowledge base when queried for evidence of an association between drug X and renal failure. Bold text indicates hypothetical hyperlinks that will take the expert directly to more detailed information
A shared experience across these efforts was that carefully and thoughtfully specifying the criteria for establishing a positive or negative drug–HOI association is a tremendous amount of work. There was a sense of dissatisfaction that each reference set was neither large enough to allow for the breadth of analyses desired, nor sufficiently impervious to post hoc criticism. Each reference set was an important contribution to their respective efforts, while at the same time insufficient to meet the broad needs of the drug safety research community. We believe that the thorough evidence-based assessment of the effects of 1,000 active ingredients across 100 HOIs while developing the OHDSI KB will lead to a more globally useful reference standard because the task will bring together medication safety practitioners and domain experts with informatics experts who possess the technical skills necessary to implement a standardized, reproducible process for structured evidence synthesis.
3 Early Progress on the KB
3.1 The Information Sources
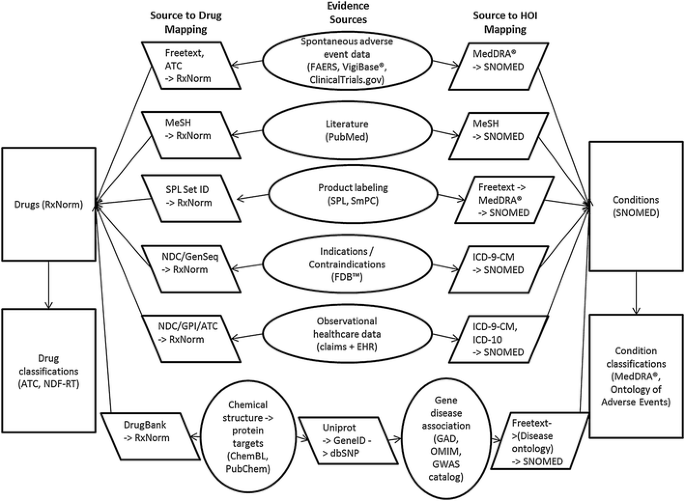
Figure 1 outlines the information sources proposed for the OHDSI KB and the necessary mappings to standardize the content across the sources. As a starting point, we have chosen RxNorm [[23](/article/10.1007/s40264-014-0189-0#ref-CR23 "Nelson SJ, Zeng K, Kilbourne J, Powell T, Moore R. Normalized names for clinical drugs: RxNorm at 6 years. J Am Med Inform Assoc. 2011;18(4):441–8. doi: 10.1136/amiajnl-2011-000116
.")\] as the standard terminology for drugs, and Systematized Nomenclature of Medicine-Clinical Terms (SNOMED-CT) \[[24](/article/10.1007/s40264-014-0189-0#ref-CR24 "Lee D, de Keizer N, Lau F, Cornet R. Literature review of SNOMED CT use. JAMIA. 2014;21(e1):e11–9. doi:
10.1136/amiajnl-2013-001636
.")\] as the standard terminology for conditions. This decision is motivated by prior work by OHDSI collaborators who lead the development of the OMOP common data model \[[25](/article/10.1007/s40264-014-0189-0#ref-CR25 "Overhage JM, Ryan PB, Reich CG, Hartzema AG, Stang PE. Validation of a common data model for active safety surveillance research. J Am Med Inform Assoc. 2012;19(1):54–60. doi:
10.1136/amiajnl-2011-000376
.")\] and standard vocabulary \[[26](/article/10.1007/s40264-014-0189-0#ref-CR26 "Defalco FJ, Ryan PB, Soledad Cepeda M. Applying standardized drug terminologies to observational healthcare databases: a case study on opioid exposure. Health Serv Outcomes Res Methodol. 2013;13(1):58–67. doi:
10.1007/s10742-012-0102-1
.")\]. The vocabulary provides mappings from RxNorm to various drug classification systems such as the Enhanced Therapeutic Classification maintained by First Databank (FDB™), the World Health Organization (WHO) Anatomical Therapeutic Chemical Classification System (ATC), and the Veteran’s Administration National Drug File-Reference Terminology (NDF-RT) \[[26](/article/10.1007/s40264-014-0189-0#ref-CR26 "Defalco FJ, Ryan PB, Soledad Cepeda M. Applying standardized drug terminologies to observational healthcare databases: a case study on opioid exposure. Health Serv Outcomes Res Methodol. 2013;13(1):58–67. doi:
10.1007/s10742-012-0102-1
.")\]. That vocabulary also contains mappings from various sources of diagnosis terminologies, such as the International Classification of Diseases, Revision 9 (ICD-9) and Revision 10 (ICD-10), into SNOMED-CT and from SNOMED-CT conditions to Medical Dictionary for Regulatory Activities (MedDRA®). We will build on previous work to extend the vocabulary to link RxNorm to DrugBank \[[27](/article/10.1007/s40264-014-0189-0#ref-CR27 "Law V, Knox C, Djoumbou Y, Jewison T, Guo AC, Liu Y, et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 2014;42(1):D1091–7. doi:
10.1093/nar/gkt1068
.")\]. This will allow for “snowball” integration of mappings from RxNorm to chemicals and protein targets (ChEMBL and PubChem), genes (UniProt), gene–disease associations in other National Center for Biotechnology Information databases, and back to SNOMED-CT via Disease Ontology \[[28](/article/10.1007/s40264-014-0189-0#ref-CR28 "Schriml LM, Arze C, Nadendla S, Chang YW, Mazaitis M, Felix V, et al. Disease ontology: a backbone for disease semantic integration. Nucleic Acids Res. 2012;40(Database issue):D940–6. doi:
10.1093/nar/gkr972
.")\].Fig. 1
Information sources proposed for the initial version of the OHDSI knowledge base. ATC Anatomical Therapeutic Chemical Classification System, EHR electronic health record, FAERS Federal Drug Administration Adverse Event Reporting System, FDB™ First DataBank, GAD Genetic Association Database, GPI Generic Product Identifier, GWAS Genome-wide association study, HOI health outcome of interest, ICD-10 International Classification of Diseases, Tenth Revision, ICD-9-CM International Classification of Diseases, Ninth Revision, Clinical Modification, MeSH Medical Subject Headings, NDC National Drug Code Directory, NDF-RT National Drug File-Reference Terminology, OHDSI Observational Health Data Sciences and Informatics, OMIM Online Mendelian Inheritance in Man, SmPC EU Summary of Product Characteristics, SNOMED Systematized Nomenclature of Medicine, SPL Structured Produce Labeling
Other sources shown in Fig. 1 include spontaneous adverse event reporting data from the US Food and Drug Administration (FDA) Adverse Event Reporting System (FAERS) and WHO VigiBase®, which allows for disproportionality analysis. Additional information on adverse events will come from the ClinicalTrials.gov clinical trials registry [[29](/article/10.1007/s40264-014-0189-0#ref-CR29 "Zarin DA, Tse T, Williams RJ, Califf RM, Ide NC. The ClinicalTrials.gov results database—update and key issues. N Engl J Med. 2011;364(9):852–60. doi: 10.1056/NEJMsa1012065
.")\], which now links adverse events reported during clinical trials to important intervention and study design information. A subset of PubMed will be filtered as described above, and the KB will provide links from Medical Subject Headings (MeSH) concepts to RxNorm drugs and SNOMED-CT conditions. US Structured Product Labeling (SPL) contains tagged entities for drug active ingredients that the KB will link to RxNorm drugs. Also, we will use a text mining tool called SPLICER to extract adverse event information present in the boxed warnings, warning/precaution, and adverse reaction sections of SPLs, and link the extracted information to RxNorm drugs and SNOMED-CT conditions \[[30](/article/10.1007/s40264-014-0189-0#ref-CR30 "Duke J, Friedlin J, Ryan P. A quantitative analysis of adverse events and “overwarning” in drug labeling. Arch Intern Med. 2011;171(10):944–6. doi:
10.1001/archinternmed.2011.182
."), [31](/article/10.1007/s40264-014-0189-0#ref-CR31 "Duke JD, Friedlin J. ADESSA: a real-time decision support service for delivery of semantically coded adverse drug event data. AMIA Annu Symp Proc. 2010;2010:177–81.")\]. The KB will also include drug–HOI association data derived from observational healthcare datasets, using methods developed during the OMOP and EU-ADR efforts \[[6](/article/10.1007/s40264-014-0189-0#ref-CR6 "DuMouchel W, Ryan PB, Schuemie MJ, Madigan D. Evaluation of disproportionality safety signaling applied to healthcare databases. Drug Saf. 2013;36(Suppl 1):S123–32. doi:
10.1007/s40264-013-0106-y
.")–[16](/article/10.1007/s40264-014-0189-0#ref-CR16 "Schuemie MJ, Gini R, Coloma PM, Straatman H, Herings RM, Pedersen L, et al. Replication of the OMOP experiment in Europe: evaluating methods for risk identification in electronic health record databases. Drug Saf. 2013;36(Suppl 1):S159–69. doi:
10.1007/s40264-013-0109-8
.")\].3.2 Iterative Development of the Reference Standard, Incremental Extensions to the KB
To be successful, the KB has to make it simpler for practitioners to access, retrieve, and synthesize evidence so that they can reach a rigorous and accurate assessment of causal relationships between a given drug and HOI. Given a potential causal relationship, there might be a need to assess causality at the individual case level or at the “global” level that considers the overall body evidence. In individual cases, a number of structured decision processes have been proposed since the 1970s [32], ranging from simple psychometrically weighted questionnaires [33–36] to probabilistic algorithms that calculate the probability in favor of a drug–HOI association on the basis of epidemiological and patient case information [[37](/article/10.1007/s40264-014-0189-0#ref-CR37 "Koh Y, Yap CW, Li SC. A quantitative approach of using genetic algorithm in designing a probability scoring system of an adverse drug reaction assessment system. Int J Med Inform. 2008;77(6):421–30. doi: 10.1016/j.ijmedinf.2007.08.010
."), [38](/article/10.1007/s40264-014-0189-0#ref-CR38 "Lanctot KL, Naranjo CA. Comparison of the Bayesian approach and a simple algorithm for assessment of adverse drug events. Clin Pharmacol Therap. 1995;58(6):692–8. doi:
10.1016/0009-9236(95)90026-8
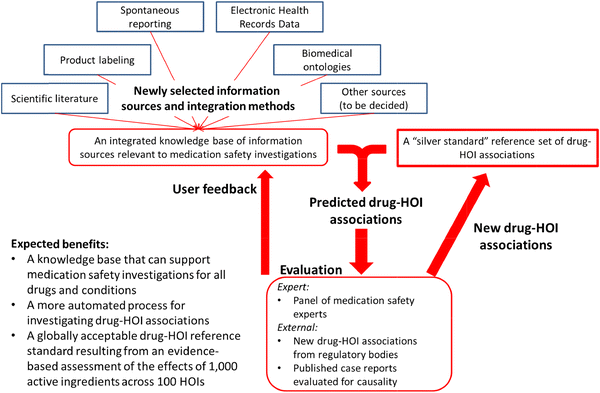
.")\]. Our task is not to judge between these processes, but to help practitioners more efficiently gather together information that would help them use the process they deem most appropriate for a given task (e.g., prior reports and the prevalence of events in exposed and non-exposed patients). Practitioners assessing the total body of evidence for a drug–HOI association would benefit from the KB’s comprehensive inclusion of evidence sources and its ability to query across all of the sources, using a small set of standardized vocabularies.Figure 2 shows the iterative process we plan to use to accomplish these goals. The OHDSI team will select an initial set of data sources and integrate them into a common format. All content in this initial version of the KB will be time-stamped for when it was generated (e.g., the date when relevant case reports, observational studies and randomized controlled trials were published in scientific journals, when disproportionality analysis met signaling thresholds in spontaneous reporting systems, and when adverse events were added to product labeling). It is also important to note that the KB will include evidence items that report no finding of a causal association between a drug and HOI so that experts will be able to gather information from all relevant sources.
Fig. 2
A systems view of OHDSI knowledge base development. HOI health outcome of interest, OHDSI Observational Health Data Sciences and Informatics
An important goal of this project is to develop a more automated process for establishing positive and negative control drug–HOI associations. Toward that end, a panel of drug safety experts will use the first version of the KB to review existing reference sets (Table 2) and establish an initial “silver” standard of drug–HOI associations that the panel finds credible with a high level of inter-rater agreement. This “silver” standard will serve as the basis for training a classification model, which will take as inputs features (“covariates”) derived from the KB and output predicted positive and negative drug–HOI associations. We will also see if the model is able to predict any associations identified by regulatory bodies or published case reports that the panel reviews after initial construction of the KB. Iterative versions of the model will be developed as the expert panel proceeds to evaluate drug–HOI combinations from the 1,000 × 100 matrix.
The process described above, and shown in Fig. 2, will also help identify changes that will enhance the usability of the KB for future users. At the same time, an error analysis of the prediction algorithm will help us to identify necessary modifications to the information sources or integration methods that might improve prediction accuracy. This entire procedure will be repeated, iteratively expanding the “silver” standard and improving the KB, until the expert panel accomplishes the evidence assessment goal. The result will be a reference standard covering the 1,000 × 100 matrix and a predictive model (or family of models) that accurately classifies whether a given drug is related to an HOI, on the basis of the available evidence from all sources (Fig. 3). High-performance models might ultimately provide a probabilistic evidence-based assessment for all drug–HOI pairs.
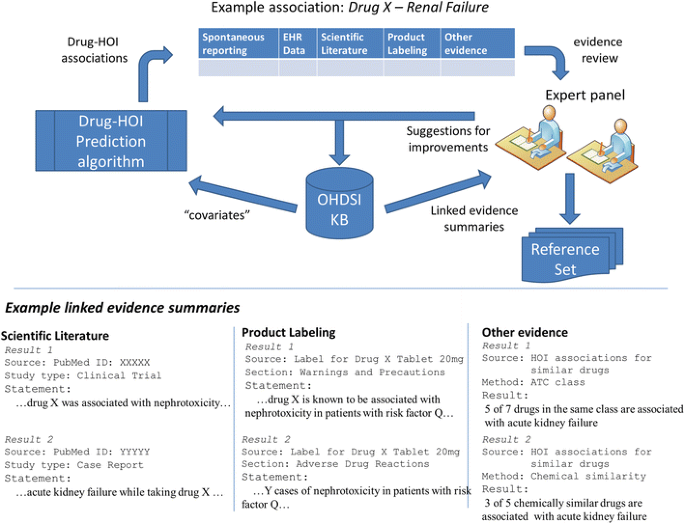
Fig. 3
Expert users will drive both the content of the knowledge base and provide feedback that will help improve the drug–HOI prediction algorithm. In this hypothetical example, the experts are able to “drill down” to review important information on various evidence items present in the KB that support an association between drug X and renal failure. ATC Anatomical Therapeutic Chemical Classification System, EHR electronic health record, HOI health outcome of interest, KB knowledge base, OHDSI Observational Health Data Sciences and Informatics
As the KB matures, we will explore the value of including innovative sources of drug safety evidence, such as inferences derived from biomedical ontologies and predictive methods emerging from the nascent field of network medicine [[4](/article/10.1007/s40264-014-0189-0#ref-CR4 "Jacunski A, Tatonetti NP. Connecting the dots: applications of network medicine in pharmacology and disease. Clin Pharmacol Therap. 2013;94(6):659–69. doi: 10.1038/clpt.2013.168
.")\]. A number of new methods are worth considering, including Duke et al.’s \[[39](/article/10.1007/s40264-014-0189-0#ref-CR39 "Duke JD, Han X, Wang ZP, Subhadarshini A, Karnik SD, Li XC et al. Literature based drug interaction prediction with clinical assessment using electronic medical records: novel myopathy associated drug interactions. Plos Comput Biol. 2012;8(8):e1002614. doi:
10.1371/journal.pcbi.1002614
.")\] template-based approach to inferring drug-interaction predictions using metabolic pathways extracted from the scientific literature, models that infer adverse events from graphical models of drug and conditions \[[40](/article/10.1007/s40264-014-0189-0#ref-CR40 "Cami A, Arnold A, Manzi S, Reis B. Predicting adverse drug events using pharmacological network models. Sci Transl Med. 2011;3(114):114ra127. doi:
10.1126/scitranslmed.3002774
.")–[42](/article/10.1007/s40264-014-0189-0#ref-CR42 "Cheng FX, Li WH, Wang XC, Zhou YD, Wu ZR, Shen J, et al. Adverse drug events: database construction and in silico prediction. J Chem Inf Model. 2013;53(4):744–52. doi:
10.1021/Ci4000079
.")\], and methods that use innovative approaches to overcome known limitations of drug safety sources such as spontaneous adverse event reports \[[18](/article/10.1007/s40264-014-0189-0#ref-CR18 "Tatonetti NP, Ye PP, Daneshjou R, Altman RB. Data-driven prediction of drug effects and interactions. Sci Transl Med. 2012;4(125):125ra31 doi:
10.1126/scitranslmed.3003377
.")\] and electronic healthcare databases \[[43](/article/10.1007/s40264-014-0189-0#ref-CR43 "Harpaz R, DuMouchel W, Shah NH, Madigan D, Ryan P, Friedman C. Novel data-mining methodologies for adverse drug event discovery and analysis. Clin Pharmacol Therap. 2012;91(6):1010–21. doi:
10.1038/clpt.2012.50
.")\]. As each information source is brought into the KB, we will empirically assess its added value in classifying drug–outcome pairs. By tying the quality and coverage of the KB to explicit performance characteristics, we will know if an addition to the KB moves us toward or away from a more systematically informed scientific process.4 A Hypothetical Example of Using the OHDSI KB
Here, we provide a hypothetical example of how the KB might be used to reconcile of disparate sources of evidence relevant to assessing a drug–HOI association. Imagine that an expert is investigating the possible association of some active ingredient (Drug X) with kidney injury. The expert would query the KB using the RxNorm identifier for Drug X and the SNOMED term Renal failure syndrome (disorder). Results from this hypothetical query are shown in Table 3. The first columns show some basic information, including that there is no known contraindication between the drug and HOI. The remaining columns show the sources of evidence available in the KB with additional information including:
- whether the HOI is mentioned as an adverse drug reaction in product labeling and when it first appeared in each source,
- the number of studies indexed in the scientific literatures in which drug and HOI terms co-occur,
- whether pharmacovigilance signals have been identified from spontaneous reporting, which datasets, and when,
- whether pharmacovigilance signals have been identified in electronic health records data, which datasets, and when
After reviewing this initial summary of the evidence available in the KB, the expert can “drill down” to examine relevant details. Underlined text in Table 3 indicates hyperlinks that will take the expert directly to more detailed information. Figure 3 shows that the specific information that the KB will provide is driven by expert users as we develop the KB.
5 Summary and Conclusions
We believe that striving toward the goal of a generally useful KB, though ambitious, is necessary for advancing the science of drug safety. Individually, each data source is insufficient to provide the evidence required for a reliable inference in the general case and a reference set in our specific case. Spontaneous adverse event reporting data remains a foundational component of drug safety, but well-acknowledged limitations of underreporting and lack of an available denominator make analysis of these data subject to various sources of bias [[2](/article/10.1007/s40264-014-0189-0#ref-CR2 "Yu AC. Methods in biomedical ontology. J Biomed Inform. 2006;39(3):252–66. doi: 10.1016/j.jbi.2005.11.006
."), [3](/article/10.1007/s40264-014-0189-0#ref-CR3 "Marshall MS, Boyce R, Deus HF, Zhao J, Willighagen EL, Samwald M, et al. Emerging practices for mapping and linking life sciences data using RDF—a case series. Web Semant Sci Serv Agents World Wide Web. 2012;14:2–13."), [26](/article/10.1007/s40264-014-0189-0#ref-CR26 "Defalco FJ, Ryan PB, Soledad Cepeda M. Applying standardized drug terminologies to observational healthcare databases: a case study on opioid exposure. Health Serv Outcomes Res Methodol. 2013;13(1):58–67. doi:
10.1007/s10742-012-0102-1
."), [29](/article/10.1007/s40264-014-0189-0#ref-CR29 "Zarin DA, Tse T, Williams RJ, Califf RM, Ide NC. The ClinicalTrials.gov results database—update and key issues. N Engl J Med. 2011;364(9):852–60. doi:
10.1056/NEJMsa1012065
."), [44](/article/10.1007/s40264-014-0189-0#ref-CR44 "Juhlin K, Ye X, Star K, Norén GN. Outlier removal to uncover patterns in adverse drug reaction surveillance—a simple unmasking strategy. Pharmacoepidemiol Drug Saf. 2013;22(10):1119–29. doi:
10.002/pds.3474
."), [45](/article/10.1007/s40264-014-0189-0#ref-CR45 "Karimi G, Star K, Norén GN, Hagg S. The impact of duration of treatment on reported time-to-onset in spontaneous reporting systems for pharmacovigilance. PLoS One. 2013;8(7):e68938. doi:
10.1371/journal.pone.0068938
.")\]. Product labeling serves as a primary source of information collected during the clinical development program, but primarily originates from clinical trials that are often underpowered for detecting rare adverse events, have insufficient follow-up for long-term adverse events, and comprise patient populations who may not be representative of the patients exposed to the drug in the real world. The level of confidence that adverse event information is credible versus “overwarning” can vary on the basis of whether it is mentioned in the boxed warning, precautions, or adverse reactions sections. Moreover, it is often the case that only limited supporting data are available to quantify the risk of a mentioned adverse event, and products can have multiple labels with inconsistent safety information \[[30](/article/10.1007/s40264-014-0189-0#ref-CR30 "Duke J, Friedlin J, Ryan P. A quantitative analysis of adverse events and “overwarning” in drug labeling. Arch Intern Med. 2011;171(10):944–6. doi:
10.1001/archinternmed.2011.182
."), [46](/article/10.1007/s40264-014-0189-0#ref-CR46 "Duke J, Friedlin J, Li X. Consistency in the safety labeling of bioequivalent medications. Pharmacoepidemiol Drug Saf. 2013;22(3):294–301. doi:
10.1002/pds.3351
.")\]. Observational databases often offer the largest source for patient-level data with real-world experience, but epidemiological studies are often challenged by confounding and other sources of bias that threaten the validity of results. While each contributing data source has substantial limitations, we believe that these can be substantially mitigated by the KB development approach that we propose.In addition to generating the KB, we also plan to work toward an efficient automated process for regular maintenance and revision. Currency of information is of considerable interest in drug safety, as product manufacturers and regulatory agencies strive to identify drug-related adverse events as soon as possible during the lifecycle of the product, and providers and patients expect that their medical decision-making can be informed by the most reliable and timely evidence available. The systematic upkeep of the KB will not only preserve relative consistency between the original sources and the composite summary as knowledge evolves over time, but might also facilitate more efficient evidence dissemination across all interested stakeholders.
To be sustainable, the KB requires an open-source, community-led effort that complements the other existing business models to offer the entire community a more complete solution to the problem. By bringing together pharmacovigilance and informatics experts into an open collaboration, we expect feedback from stakeholders that will help identify missing information, sources that should be added to the KB, and corrections or modifications to the sources represented in the KB. Persons interested in become collaborators can contact us directly or through the OHDSI project management site (http://goo.gl/TRSUoH) or the OHDSI code development sites (https://github.com/OHDSI/KnowledgeBase).
In conclusion, we are excited to help jumpstart this community effort, as we fully expect a drug safety KB will become an invaluable tool for methodological research and pharmacovigilance practice alike.
References
- Bush V. As we may think. The Atlantic. July 1945.
- Yu AC. Methods in biomedical ontology. J Biomed Inform. 2006;39(3):252–66. doi:10.1016/j.jbi.2005.11.006.
Article PubMed Google Scholar - Marshall MS, Boyce R, Deus HF, Zhao J, Willighagen EL, Samwald M, et al. Emerging practices for mapping and linking life sciences data using RDF—a case series. Web Semant Sci Serv Agents World Wide Web. 2012;14:2–13.
Article Google Scholar - Jacunski A, Tatonetti NP. Connecting the dots: applications of network medicine in pharmacology and disease. Clin Pharmacol Therap. 2013;94(6):659–69. doi:10.1038/clpt.2013.168.
Article CAS Google Scholar - Yeleswarapu S, Rao A, Joseph T, Saipradeep VG, Srinivasan R. A pipeline to extract drug-adverse event pairs from multiple data sources. BMC Med Inform Decis Mak. 2014;14(1):13.
Article PubMed Central PubMed Google Scholar - DuMouchel W, Ryan PB, Schuemie MJ, Madigan D. Evaluation of disproportionality safety signaling applied to healthcare databases. Drug Saf. 2013;36(Suppl 1):S123–32. doi:10.1007/s40264-013-0106-y.
Article PubMed Google Scholar - Madigan D, Schuemie MJ, Ryan PB. Empirical performance of the case-control method: lessons for developing a risk identification and analysis system. Drug Saf. 2013;36(Suppl 1):S73–82. doi:10.1007/s40264-013-0105-z.
Article PubMed Google Scholar - Ryan PB, Madigan D, Stang PE, Overhage JM, Racoosin JA, Hartzema AG. Empirical assessment of methods for risk identification in healthcare data: results from the experiments of the Observational Medical Outcomes Partnership. Stat Med. 2012;31(30):4401–15. doi:10.1002/sim.5620.
Article PubMed Google Scholar - Ryan PB, Schuemie MJ. Evaluating performance of risk identification methods through a large-scale simulation of observational data. Drug Saf. 2013;36(Suppl 1):S171–80. doi:10.1007/s40264-013-0110-2.
Article PubMed Google Scholar - Ryan PB, Schuemie MJ, Gruber S, Zorych I, Madigan D. Empirical performance of a new user cohort method: lessons for developing a risk identification and analysis system. Drug Saf. 2013;36(Suppl 1):S59–72. doi:10.1007/s40264-013-0099-6.
Article PubMed Google Scholar - Ryan PB, Schuemie MJ, Madigan D. Empirical performance of a self-controlled cohort method: lessons for developing a risk identification and analysis system. Drug Saf. 2013;36(Suppl 1):S95–106. doi:10.1007/s40264-013-0101-3.
Article PubMed Google Scholar - Ryan PB, Stang PE, Overhage JM, Suchard MA, Hartzema AG, DuMouchel W, et al. A comparison of the empirical performance of methods for a risk identification system. Drug Saf. 2013;36(Suppl 1):S143–58. doi:10.1007/s40264-013-0108-9.
Article PubMed Google Scholar - Schuemie MJ, Madigan D, Ryan PB. Empirical performance of LGPS and LEOPARD: lessons for developing a risk identification and analysis system. Drug Saf. 2013;36(Suppl 1):S133–42. doi:10.1007/s40264-013-0107-x.
Article PubMed Google Scholar - Suchard MA, Zorych I, Simpson SE, Schuemie MJ, Ryan PB, Madigan D. Empirical performance of the self-controlled case series design: lessons for developing a risk identification and analysis system. Drug Saf. 2013;36(Suppl 1):S83–93. doi:10.1007/s40264-013-0100-4.
Article PubMed Google Scholar - Schuemie MJ, Coloma PM, Straatman H, Herings RM, Trifiro G, Matthews JN, et al. Using electronic health care records for drug safety signal detection: a comparative evaluation of statistical methods. Med Care. 2012;50(10):890–7.
Article PubMed Google Scholar - Schuemie MJ, Gini R, Coloma PM, Straatman H, Herings RM, Pedersen L, et al. Replication of the OMOP experiment in Europe: evaluating methods for risk identification in electronic health record databases. Drug Saf. 2013;36(Suppl 1):S159–69. doi:10.1007/s40264-013-0109-8.
Article PubMed Google Scholar - Alvarez Y, Hidalgo A, Maignen F, Slattery J. Validation of statistical signal detection procedures in eudravigilance post-authorization data: a retrospective evaluation of the potential for earlier signalling. Drug Saf. 2010;33(6):475–87. doi:10.2165/11534410-000000000-00000.
Article PubMed Google Scholar - Tatonetti NP, Ye PP, Daneshjou R, Altman RB. Data-driven prediction of drug effects and interactions. Sci Transl Med. 2012;4(125):125ra31 doi:10.1126/scitranslmed.3003377.
Article PubMed Central PubMed Google Scholar - Hochberg AM, Hauben M, Pearson RK, O’Hara DJ, Reisinger SJ, Goldsmith DI, et al. An evaluation of three signal-detection algorithms using a highly inclusive reference event database. Drug Saf. 2009;32(6):509–25. doi:10.2165/00002018-200932060-00007.
Article PubMed Google Scholar - Norén GN, Bergvall T, Ryan PB, Juhlin K, Schuemie MJ, Madigan D. Empirical performance of the calibrated self-controlled cohort analysis within temporal pattern discovery: lessons for developing a risk identification and analysis system. Drug Saf. 2013;36(Suppl 1):S107–21. doi:10.1007/s40264-013-0095-x.
Article PubMed Google Scholar - Ryan PB, Schuemie MJ, Welebob E, Duke J, Valentine S, Hartzema AG. Defining a reference set to support methodological research in drug safety. Drug Saf. 2013;36(Suppl 1):S33–47. doi:10.1007/s40264-013-0097-8.
Article PubMed Google Scholar - Coloma PM, Avillach P, Salvo F, Schuemie MJ, Ferrajolo C, Pariente A, et al. A reference standard for evaluation of methods for drug safety signal detection using electronic healthcare record databases. Drug Saf. 2013;36(1):13–23. doi:10.1007/s40264-012-0002-x.
Article CAS PubMed Google Scholar - Nelson SJ, Zeng K, Kilbourne J, Powell T, Moore R. Normalized names for clinical drugs: RxNorm at 6 years. J Am Med Inform Assoc. 2011;18(4):441–8. doi:10.1136/amiajnl-2011-000116.
PubMed Central PubMed Google Scholar - Lee D, de Keizer N, Lau F, Cornet R. Literature review of SNOMED CT use. JAMIA. 2014;21(e1):e11–9. doi:10.1136/amiajnl-2013-001636.
PubMed Google Scholar - Overhage JM, Ryan PB, Reich CG, Hartzema AG, Stang PE. Validation of a common data model for active safety surveillance research. J Am Med Inform Assoc. 2012;19(1):54–60. doi:10.1136/amiajnl-2011-000376.
PubMed Central PubMed Google Scholar - Defalco FJ, Ryan PB, Soledad Cepeda M. Applying standardized drug terminologies to observational healthcare databases: a case study on opioid exposure. Health Serv Outcomes Res Methodol. 2013;13(1):58–67. doi:10.1007/s10742-012-0102-1.
Article PubMed Central PubMed Google Scholar - Law V, Knox C, Djoumbou Y, Jewison T, Guo AC, Liu Y, et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 2014;42(1):D1091–7. doi:10.1093/nar/gkt1068.
Article CAS PubMed Central PubMed Google Scholar - Schriml LM, Arze C, Nadendla S, Chang YW, Mazaitis M, Felix V, et al. Disease ontology: a backbone for disease semantic integration. Nucleic Acids Res. 2012;40(Database issue):D940–6. doi:10.1093/nar/gkr972.
Article CAS PubMed Central PubMed Google Scholar - Zarin DA, Tse T, Williams RJ, Califf RM, Ide NC. The ClinicalTrials.gov results database—update and key issues. N Engl J Med. 2011;364(9):852–60. doi:10.1056/NEJMsa1012065.
Article CAS PubMed Central PubMed Google Scholar - Duke J, Friedlin J, Ryan P. A quantitative analysis of adverse events and “overwarning” in drug labeling. Arch Intern Med. 2011;171(10):944–6. doi:10.1001/archinternmed.2011.182.
Article PubMed Google Scholar - Duke JD, Friedlin J. ADESSA: a real-time decision support service for delivery of semantically coded adverse drug event data. AMIA Annu Symp Proc. 2010;2010:177–81.
PubMed Central PubMed Google Scholar - Agbabiaka TB, Savovic J, Ernst E. Methods for causality assessment of adverse drug reactions: a systematic review. Drug Saf. 2008;31(1):21–37.
Article PubMed Google Scholar - Karch FE, Lasagna L. Toward the operational identification of adverse drug reactions. Clin Pharmacol Therap. 1977;21(3):247–54.
CAS Google Scholar - Karch FE, Smith CL, Kerzner B, Mazzullo JM, Weintraub M, Lasagna L. Adverse drug reactions—a matter of opinion. Clin Pharmacol Therap. 1976;19(5 Pt 1):489–92.
CAS Google Scholar - Koh Y, Li SC. A new algorithm to identify the causality of adverse drug reactions. Drug Saf. 2005;28(12):1159–61.
Article PubMed Google Scholar - Naranjo CA, Busto U, Sellers EM, Sandor P, Ruiz I, Roberts EA, et al. A method for estimating the probability of adverse drug-reactions. Clin Pharmacol Therap. 1981;30(2):239–45.
Article CAS Google Scholar - Koh Y, Yap CW, Li SC. A quantitative approach of using genetic algorithm in designing a probability scoring system of an adverse drug reaction assessment system. Int J Med Inform. 2008;77(6):421–30. doi:10.1016/j.ijmedinf.2007.08.010.
Article PubMed Google Scholar - Lanctot KL, Naranjo CA. Comparison of the Bayesian approach and a simple algorithm for assessment of adverse drug events. Clin Pharmacol Therap. 1995;58(6):692–8. doi:10.1016/0009-9236(95)90026-8.
Article CAS Google Scholar - Duke JD, Han X, Wang ZP, Subhadarshini A, Karnik SD, Li XC et al. Literature based drug interaction prediction with clinical assessment using electronic medical records: novel myopathy associated drug interactions. Plos Comput Biol. 2012;8(8):e1002614. doi:10.1371/journal.pcbi.1002614.
Article CAS PubMed Central PubMed Google Scholar - Cami A, Arnold A, Manzi S, Reis B. Predicting adverse drug events using pharmacological network models. Sci Transl Med. 2011;3(114):114ra127. doi:10.1126/scitranslmed.3002774.
Article PubMed Google Scholar - Cami A, Manzi S, Arnold A, Reis BY. Pharmacointeraction network models predict unknown drug–drug interactions. Plos One. 2013;8(4):e61468. doi:10.1371/journal.pone.0061468.
Article CAS PubMed Central PubMed Google Scholar - Cheng FX, Li WH, Wang XC, Zhou YD, Wu ZR, Shen J, et al. Adverse drug events: database construction and in silico prediction. J Chem Inf Model. 2013;53(4):744–52. doi:10.1021/Ci4000079.
Article CAS PubMed Google Scholar - Harpaz R, DuMouchel W, Shah NH, Madigan D, Ryan P, Friedman C. Novel data-mining methodologies for adverse drug event discovery and analysis. Clin Pharmacol Therap. 2012;91(6):1010–21. doi:10.1038/clpt.2012.50.
Article CAS Google Scholar - Juhlin K, Ye X, Star K, Norén GN. Outlier removal to uncover patterns in adverse drug reaction surveillance—a simple unmasking strategy. Pharmacoepidemiol Drug Saf. 2013;22(10):1119–29. doi:10.002/pds.3474.
PubMed Google Scholar - Karimi G, Star K, Norén GN, Hagg S. The impact of duration of treatment on reported time-to-onset in spontaneous reporting systems for pharmacovigilance. PLoS One. 2013;8(7):e68938. doi:10.1371/journal.pone.0068938.
Article CAS PubMed Central PubMed Google Scholar - Duke J, Friedlin J, Li X. Consistency in the safety labeling of bioequivalent medications. Pharmacoepidemiol Drug Saf. 2013;22(3):294–301. doi:10.1002/pds.3351.
Article CAS PubMed Google Scholar
Funding Support
First author (Richard Boyce) is funded by National Institute on Aging grant K01AG044433 and National Library of Medicine grant 1R01LM011838-01. Vojtech Huser is supported by the Intramural Research Program of the National Institutes of Health Clinical Center and the National Library of Medicine.
Patrick Ryan, Martijn Schuemie, and Erica Voss are employees of Janssen Research and Development. Christian Reich is an employee of AstraZeneca. Abraham Hartzema received funding from Pfizer, although not for this project; he is also a paid senior consultant to the FDA CDRH; the content in this manuscript reflects his own opinion and not that of the FDA. Rave Harpaz is an employee of Oracle. Jon Duke has received research funding from pharmaceutical industry sources, including Merck, Janssen and Lilly.
Richard Boyce, Niklas Norén, Nicholas Tatonetti, Gianluca Trifirò, Marc Overhage, Mark Khayter, Christophe Lambert, Vojtech Huser, and Michel Dumontier have no conflicts of interest that are directly relevant to the content of this article.
Author information
Authors and Affiliations
- University of Pittsburgh, Pittsburgh, PA, USA
Richard D. Boyce - Janssen Research and Development, Titusville, NJ, USA
Patrick B. Ryan, Martijn J. Schuemie & Erica A. Voss - Uppsala Monitoring Centre, Uppsala, Sweden
G. Niklas Norén - AstraZeneca, Waltham, MA, USA
Christian Reich - Regenstrief Institute, Indianapolis, IN, USA
Jon Duke - Columbia University, New York, NY, USA
Nicholas P. Tatonetti - University of Messina, Messina, Italy
Gianluca Trifirò - Erasmus University Medical Center, Rotterdam, The Netherlands
Gianluca Trifirò - Stanford University, Palo Alto, CA, USA
Rave Harpaz - Siemens Healthcare, Malvern, PA, USA
J. Marc Overhage - University of Florida, Gainesville, FL, USA
Abraham G. Hartzema - Ephir, Boston, MA, USA
Mark Khayter - Montana State University, Bozeman, MT, USA
Christophe G. Lambert - National Institutes of Health, Bethesda, MD, USA
Vojtech Huser - Stanford University, Stanford, CA, USA
Michel Dumontier
Authors
- Richard D. Boyce
You can also search for this author inPubMed Google Scholar - Patrick B. Ryan
You can also search for this author inPubMed Google Scholar - G. Niklas Norén
You can also search for this author inPubMed Google Scholar - Martijn J. Schuemie
You can also search for this author inPubMed Google Scholar - Christian Reich
You can also search for this author inPubMed Google Scholar - Jon Duke
You can also search for this author inPubMed Google Scholar - Nicholas P. Tatonetti
You can also search for this author inPubMed Google Scholar - Gianluca Trifirò
You can also search for this author inPubMed Google Scholar - Rave Harpaz
You can also search for this author inPubMed Google Scholar - J. Marc Overhage
You can also search for this author inPubMed Google Scholar - Abraham G. Hartzema
You can also search for this author inPubMed Google Scholar - Mark Khayter
You can also search for this author inPubMed Google Scholar - Erica A. Voss
You can also search for this author inPubMed Google Scholar - Christophe G. Lambert
You can also search for this author inPubMed Google Scholar - Vojtech Huser
You can also search for this author inPubMed Google Scholar - Michel Dumontier
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toRichard D. Boyce.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Boyce, R.D., Ryan, P.B., Norén, G.N. et al. Bridging Islands of Information to Establish an Integrated Knowledge Base of Drugs and Health Outcomes of Interest.Drug Saf 37, 557–567 (2014). https://doi.org/10.1007/s40264-014-0189-0
- Published: 02 July 2014
- Issue Date: August 2014
- DOI: https://doi.org/10.1007/s40264-014-0189-0