Notch signalling is linked to epidermal cell differentiation level in basal cell carcinoma, psoriasis and wound healing (original) (raw)
Introduction
In mammalian epidermis, keratinocytes progress through successive phenotypic stages as they migrate from the germinative basal layer to the skin surface. These continuous proliferative and differentiative processes result in tissue homeostasis. Studies on embryonic mice and rats have shown the involvement of the Notch pathway in epidermal differentiation [1, 2] as well as cutaneous appendage patterning [3–5]. This, and recent work on the involvement of this pathway in adults [6], allows us to assess the transcription level of several Notch-related genes, depending on the terminal differentiation state of the keratinocytes.
The Notch system was first described as being responsible for neurogenesis and ectodermal specification in the fruit fly [7]. In chick and mouse embryos, it was shown to be involved in limb bud outgrowth and somitogenesis [8]. This pathway includes Notch receptors, trans-membrane ligands, Delta/Jagged, and Fringe proteins. By now, it has been demonstrated to be a cell-cell signalling implicated in several developmental events: lateral cell inhibition, cell fate determination, boundary formation and patterning [7, 8]. In vertebrates, several molecules of the pathway have been identified. These include (a) Notch 1–4 transmembrane receptors, (b) two families of transmembrane ligands with numerous EGF repeats, Delta 1–3 and Jagged 1 and 2, and (c) three proteins modulating the ligand-receptor induced signal, termed Lunatic Fringe, Radical Fringe and Manic Fringe. The complexity of the Notch ligand-receptor system suggests that a high number of combined associations is necessary for multipurpose signalling. Once the receptor is activated by a ligand from a neighbouring cell, proteolytic cleavage results in the liberation of the intracytoplasmic domain of Notch. The intracytoplasmic domain is then translocated into the nucleus, where it associates with the transcription factor RBPJk/Su(H) to activate the expression of downstream target genes [9]. This signalling is able to regulate cell-autonomously the level of Notch partners, and to determine the fate of neighbouring cells.
In many developing tissues, these signals inhibit differentiation. However, recent data show its role in cell differentiation in haematopoietic lineages, in pancreatic progenitor cell differentiation, and neural cell outgrowth [10–12]. In mammals, several mutations in Notch genes leading to severe disorders have been reported. A translocation which damages the Notch1 gene induces the expansion of undifferentiated progenitors [13, 14]. Missense mutations, which affect the Notch3 gene, were thought to alter the interactions with the Fringe proteins [15, 16]. Mutations have also been identified in mouse Notch4, which result in the absence of a regulatory action on the extra cellular domain [17] and in mammary tumorigenesis [18]. Recent work on Notch ligands confirmed that the regulatory action on the extra cellular domain was crucial in human adults for self-renewal and differentiation of progenitor cells. In humans, mutation of the Jagged1 gene induces a disorder called Alagille [19, 20]. In this pathology, truncated proteins may be secreted, resulting in a dominant negative effect on Notch signalling [21]. Ewing's sarcoma may be another human malignancy in which Notch signalling pathway is implicated, but via the inappropriate activation of _Manic Fringe_[22]. Thus, in mammals, mutations in Notch genes are associated with de-regulation of cell proliferation, differentiation and migration (for a review [23]). Furthermore, whilst our work was in progress, Lowell et al [6] recently showed that Delta-Notch interactions promote differentiation of epidermal stem cells. They used transfected keratinocytes to demonstrate that the Delta 1 ligand inhibits cell autonomously Notch 1 receptor expression and that Delta 1 has to be present on cells (in trans) different from those expressing Notch 1 to trigger Notch receptor. It is a cell-cell signalling system: clusters of stem cells expressing Delta 1 activate transit amplifying cells which do not express Delta 1 but express Notch 1.
Here, we evaluate the transcription levels for the transmembrane ligands and receptors and the Fringe molecules in two clinical contexts; basal cell carcinoma (BCC) and psoriasis, as well as in the context of experimental wound healing, was compared with adult human non-lesional skin. In BCC, the most common non-melanocytic human skin cancer [24–26], hyperplasic cell division may lead to invasion of the dermis by epidermal tissues. Although such tumours are readily treatable and usually have a good prognosis, many can become aggressive with local tissue invasion and destruction. There is some understanding of the elements relating to the development of BCC, especially concerning the deregulation of Hedgehog signalling which is now firmly established [27]. Psoriasis, which affects about 2% of the Caucasian population, is characterised by hyper proliferation and aberrant differentiation of keratinocytes [28, 29]. This disease has many features in common with wound healing [30]. However, in the latter process, the regenerative behaviour of keratinocytes is transient and the epidermis returns to its normal growth and differentiation programme [31], whereas in the former the hyper proliferative behaviour persists.
Here we show that Notch signalling is associated with normally differentiated human epidermis and that expression of associated transcripts are clearly repressed in the context of cell hyper proliferation. Furthermore, this signalling pathway can be rescued by treatment in the case of psoriasis, and reactivated when wounded epidermis is repaired, confirming its involvement in keratinocyte differentiation.
Materials and Methods
Biological samples
Human skin was obtained from a mammary reduction operation at the plastic surgery department of Grenoble hospital, France. Sixteen surgical resections of basal cell carcinoma showing various grades of proliferation and invasiveness (superficial, deep nodular and infiltrative) were provided by the dermatological departments of Villejuif and Lyon hospitals, France. Four psoriatic patients from Grenoble gave informed consent for biopsies to be taken. All had stable psoriatic plaques; three of them had not been treated by systemic drugs or phototherapy in the month preceding sampling, one of them had been treated by phototherapy. In all these patients, biopsies were performed on lesional and non-lesional skin so as to obtain the epidermis. When harvested, biopsies were frozen in liquid nitrogen and stored at -80°C until use.
Wound healing procedure
Split-thickness (0.6 mm) human skin was grafted on congenitally athymic nude mice, Swiss nu/nu (Iffa Credo, Les Oncins, France), aged 5 to 7 weeks. Mice to be grafted were anaesthetised with pentobarbital, and a circular piece of skin was removed from the lateral abdominal region. Skin samples to be grafted were cut into 1 cm diameter pieces with a punch biopsy and fitted into the prepared graft sites. Grafts were protected by a dressing pad and a cohesive extensible bandage. Two months after transplantation, animals were anaesthetised with pentobarbital and excisional wounds were made centrally through the entire thickness of the grafted human skin, using a 2 mm punch. The wounds were then left exposed to air. Seven days after the injury, the mice were euthanised and the healing human graft with surrounding mouse skin was harvested. Drs. Thélu and Favier are officially licensed by the French government to use mice for experiments (Agreement numbers: 04017, A3851601).
Section procedure and observations
Frozen tissue specimens were orientated in OCT embedding medium (Miles laboratories) and stored at -80°C. Six to ten micrometer-thick cryostat sections were cut serially through the specimen and were thaw-mounted onto pre-treated slides (Super Frost plus, Menzel Gläser, Germany), then stored at -80°C until use.
Nuclei were counter-stained during 30 min incubation in a DNA fluorochrome solution (propidium iodide 10 μg ml-1 in PBS), followed by three washes in PBS. Conjonctive tissues and propidium iodide labelled nuclei were excited (wavelength range: 330–385 nm, WU filter, Olympus), and blue and red fluorescence emission was observed simultaneously. Radiolabelled riboprobes hybridised to sections were detected with silver emulsion and displayed white dots under dark field conditions.
Probes
Clones from the human EST bank were sequenced and, when necessary, poly A+ regions were eliminated and/or were sub cloned in pBluescript SK+ vector (Stratagen). Gene name, clone identification number, position in the mRNA and length are as follows: Notch 1, 700304, (3'), 0.66 kb; Notch 2, 256416, (5'), 0.73 kb; Notch 3, 261623, (5'), 1.60 kb; Delta 1, 788147, (5'), 0.60 kb; Jagged 1, 376965, (3'), 1.10 kb; Manic Fringe, 824939, (3'), 0.58 kb; Lunatic Fringe, 40887, (3'), 0.68 kb; Radical Fringe, C-32410, (3'), 1.40 kb. Appropriate restriction sites and promoter sites (T3 and T7) were used to transcribe antisense mRNA probes. Three control sense mRNA probes were synthesised using Notch 3, Jagged 1 and Lunatic Fringe plasmids as transcription templates.
In situ hybridisation
Transcript expression was assessed by in situ hybridisation using (α 35S) CTP radiolabelled antisense riboprobes according to a previously described method [32]. The hybridised sections were coated with Kodak NTB-2 liquid emulsion to reveal radiolabelling. Cold labelling was performed using an antibody raised to DIG, conjugated to alkaline phosphatase (Roche), according to manufacturer's instructions.
Reverse transcriptase-PCR experiments
mRNAs were extracted from human adult skin using oligo d(T) micro columns, according to the manufacturer's protocol (Amersham, Pharmacia). Thirty or thirty-five amplification cycles were conducted at 50°C annealing temperature using the following specific oligonucleotides:
GAPDH: AATCCCATCACCATCTTCCA and GTCATCATATTTGGCAGGTT; Lunatic Fringe: AACAAGGTGCGTCCTGTCCACTTC, and GTGGACGGCGTTCCGCTTGTTTTC; Manic Fringe: CCGCGACGTCTATGTGGG, and CAGATCCTGGGCTTGCCA; Radical Fringe: CCCGACCTCTCGGTGACA, and GGTGGAAGCCTGTTCCCG; Notch1: CACCCAGAACTGCGTGCA, and GGCAGTCAAAGCCGTCGA; Notch2: GCTGATGCTGCCAAGCGT, and CCGGGGAAGACGATCCAT; Delta1: TGCTCCTGCTGGCTTTCG, and TTCATAGAGGCCACCGTC; Jagged1: GTGCATGCTCCAATCGGC, and GAAAGGCAGCACGATGCG.
Results
In normal adult skin, radiolabelled antisense riboprobes directed against Notch 1–3 mRNA hybridised to a complementary sequence in the epidermis. All three transcripts were more strongly detected in the basal layer of interfollicular epidermis (Fig. 1A,1B,1C'). Radiolabelling with Delta1 and Jagged 1 riboprobes produced a similar signal distribution to Notch (Fig. 1D,1E'). Fringe transcripts were all localised in the basal layer (Fig. 1G,1H,1I'). The cornified layer with no cell nuclei (as detected by propidium iodide labelling) may bind different probes, inducing artefacts due to edge and condensed keratin effects. In situ hybridisation control experiments performed with three sense mRNA probes were negative (Fig. 1F,1F' and data not shown).
Figure 1
Expression of Notch 1–3 receptors, Delta1 and Jagged1 transmembrane ligands and of the three Fringe excreted proteins in unlesional human skin. Nuclear staining with propidium iodide (A-C, D-F, G-I) and darkfield observation of in situ hybridisation (A'-C', D'-F', G'-I') on cryosections. Radiolabelled riboprobes against Notch (A'-C') and its ligand (D'-E') genes are detected in the basal layer of the epidermis and to a lower extent in suprabasal layers. Fringe gene expression is restricted to the basal layer (G'-I'). Signals detected in the upper cornified layer with no nucleated cells correspond to artefacts caused by dense keratin and edge effects. The control (F, F') represents a skin section processed with the sense Jagged1 riboprobe under the same conditions. Scale bar, 100 μm; Ma, Manic; Lu, Lunatic; Ra, Radical; d: dermis; bl: basal layer; spl: suprabasal layer; cl: cornified layer; e: epidermis.
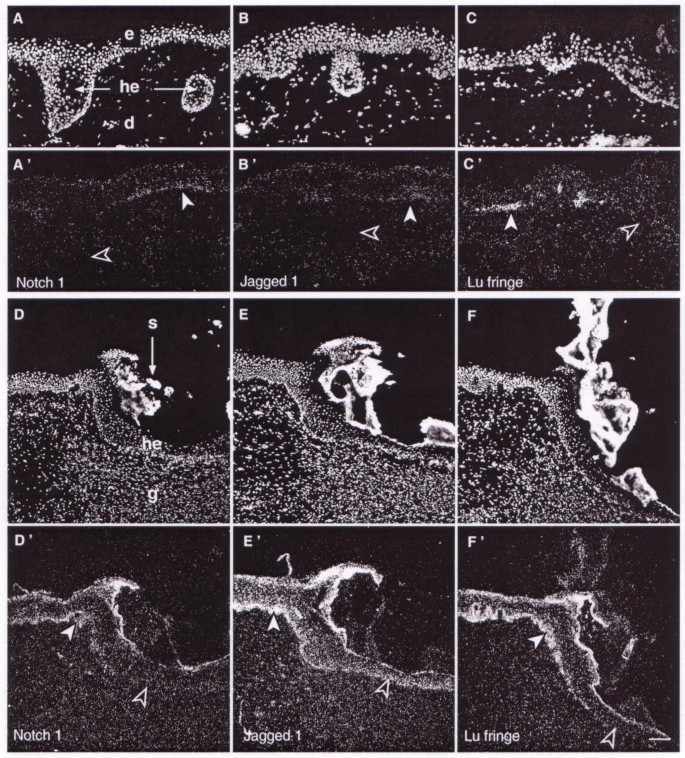
Three types of BCCs were studied: superficial, deep nodular and infiltrative. Superficial-type BCCs exhibit buds, irregular proliferation of cells attached to the epidermis and little penetration into the dermis with typical peripheral palisading cells embedded in the dermis (Fig. 2A,2B,2C and Fig. 3A). The expression of receptors, ligands and Fringe were severely lowered in these regions which display uncontrolled cell proliferation (Fig. 2A',2B',2C' and Fig. 3A). Only faint transcripts were detected in small areas, associated with almost normal epidermal structure, and conversely, no expression was observed in the palisading cells penetrating the dermis. Deep nodular BCCs, with tumour cell clusters of various size and shapes, and infiltrative type, presented no palisading peripheral cells. In these two BCC types, no expression was detected for any of the Notch-related genes tested (Fig. 3B,3C,3D,3E,3F), even when sensitive emulsion was overexposed for up to 24 days. Only small areas with normal basal layer show an expression signal.
Figure 2
Expression of Notch1 , Jagged1 and Lunatic Fringe genes in basal cell carcinoma and in experimental wound healing. Propidium iodide nuclei counterstaining (A-C, D-F) and in situ hybridisation with radiolabelled riboprobes (A'-C', D'-F'). In superficial basal cell carcinoma (A-C') Notch1, Jagged1 and Lunatic Fringe transcripts present a low level of expression in un-proliferating epidermis (filled arrowheads), and no expression in palisading cells embedded in the dermis (empty arrowheads). In wound healing experiments (D-F') gene expression in regenerated epidermis is lower in the middle (empty arrowheads) than at the wound edges (filled arrowheads). Scale bar, 100 μm; d: dermis; e: epidermis; g: granulation tissue; he: hyperproliferative epidermis; s: scab; Lu, Lunatic.
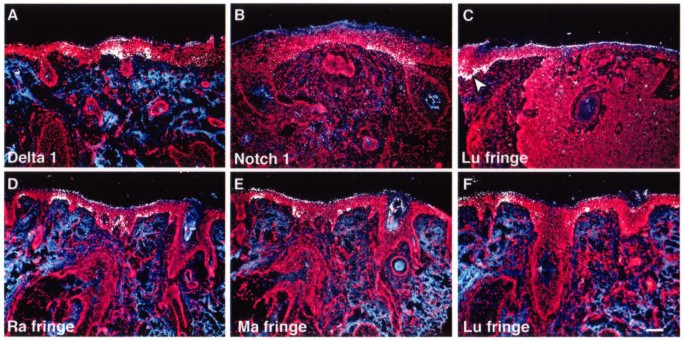
Figure 3
Expression of Delta-1 , Notch-1 and Fringe genes in four different types of basal cell carcinoma. Nuclei (red), conjonctive tissues (blue) and gene expression (white) are shown simultaneously. In superficial BCCs, only few cells express Delta-1 mRNAs in basal layer (A). The level of Notch-1 transcripts is very low in nodular BCC (B). Lu Fringe transcripts in deeply infiltrating BCC (C) are only detected at the margin of surgical excision (arrowhead; C). The expression of the three Fringe genes is restricted to small areas in basal layer in infiltrating BCC having a sclero-dermic differentiation (D-F). Scale bar, 100 μm; Ma, Manic; Lu, Lunatic; Ra, Radical.
In wound healing experiments, normal human skin was grafted on nude mice. Then, repair of experimental excisional wound [33] starts with proliferation of mouse fibroblastic cells, filling the wound space. These cells created a granulocytic tissue, which was rapidly covered with expanding keratinocytes of human epidermal origin. Proliferating cells, which are thought to be derived from the basal cell layer [33], migrated from the periphery to the centre of the wound, and progressively differentiated to constitute a pluristratified epidermis (Fig. 2D,2E,2F). Notch-related gene expression was analysed at different position points after injury in the newly regenerated basal layer. In all cases, a gradation of increasing intensity from the wound centre to the edge was evident (Fig. 2D',2E',2F'). Note also that at any given point of the wound (e.g. open arrows in Fig. 2D',2E'), the amount of Jagged1 mRNA amount is higher than that of Notch1 in the adjacent slide. This suggests that Jagged1 transcript reactivation occurs before those of Notch1, even though the time delay cannot be accurately determined under the present conditions.
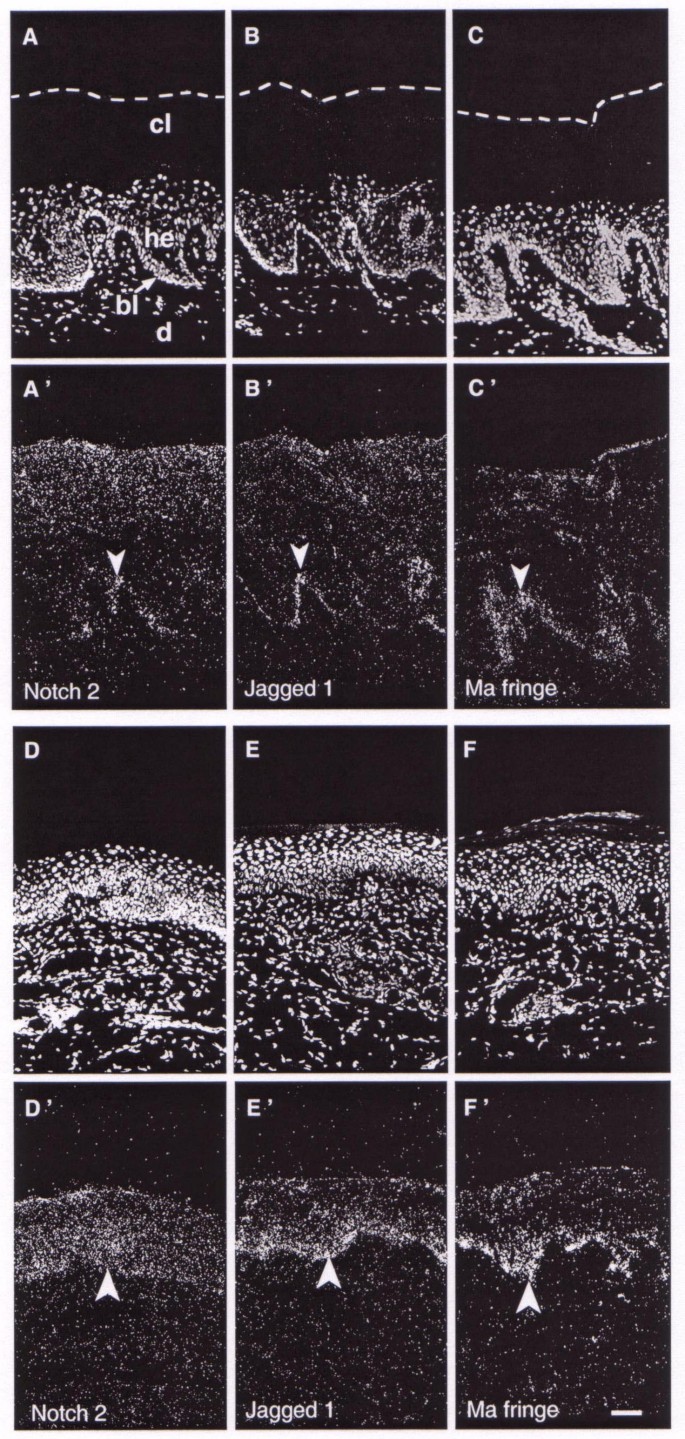
Psoriatic plaques feature hyperproliferative suprabasal layers and a thick layer of cornified cells (Fig. 4A,4B,4C). In this case, the epidermis exhibited reduced transcription of the tested genes (Fig. 4A',4B',4C'), compared with normal epidermis provided by the same individuals, which were similar to normal skin (Fig. 1). The expression in basal layers was faint in some areas and absent in others. Positive cells were located mainly at the tips of the dermal papillae (arrowheads Fig. 4A',4B',4C'). Phototherapeutic treatment of psoriatic plaques tissue reduced the hyperplasticity of the epidermis (Fig. 4D,4E,4F). In this case, hybridisation analysis showed homogeneous transcription level in the basal layer and to a relatively lesser extent in the suprabasal layers (Fig. 4D',4E',4F'). Thus, normal skin differentiation, rescued by the treatment, was linked to de novo expression of Notch, Jagged and Fringe genes, whose levels and patterns were similar to paired samples of healthy skin.
Figure 4
Expression of Notch2 , Jagged1 and Manic Fringe genes in untreated and phototherapy treated psoriatic skin. Propidium iodide nuclei counterstaining (A-C, D-F) and in situ hybridisation with radiolabelled riboprobes (A'-C', D'-F'). In psoriatic plaques (A-C'), Notch2, Jagged1 and Manic Fringe genes are down-regulated in the basal layer, and transcripts are mainly located at the tips of the dermal papillae (arrowheads). In psoriatic skin, treated by phototherapy (D-F'), gene expression levels are restored to those of normal control skin (arrowheads). Scale bar, 100 μm; Ma, Manic; d: dermis; bl: basal layer; cl: cornified layer; he: hyperproliferative epidermis.
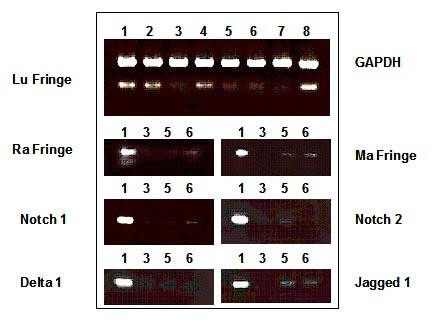
Messenger RNA integrity was controlled for by using a reverse transcriptase-polymerase chain strategy. The house-keeping GAPDH gene was present in every sample studied, as shown by the 558 bp fragment (Fig. 5, lane 1 to 8). Control samples from normal skin, unaffected skin from psoriatic patients, phototherapy treated psoriatic plaque or normal skin obtained from the margin of surgical excision expressed Lunatic Fringe gene as shown by the 309 bp band (Fig. 5, lane 1, 2, 4 and 8 respectively). However, the Lunatic Fringe 309 bp fragment was not amplified after 35 cycles in psoriatic plaque and BCC biopsies samples (Fig. 5, lane 3 and 5–7, respectively). Using a non saturating (30) number of cycle, all of the other studied genes (Radical Fringe: 300 bp, Manic Fringe: 622 bp, Notch1: 724 bp, Notch2: 473 bp, Delta1: 301 bp, Jagged1: 509 bp) were examined by RT-PCR in four representative samples: normal skin as positive control, psoriatic skin, infiltrating BCC and a nodular BCC as negative control. The results are consistent with in situ hybrisation with weak amplification being related to weak positive areas in pathological samples
Figure 5
Notch -related gene expression is down-regulated in psoriatic and in BCC skin biopsies. Using RT-PCR, the amplification rate of a 309 bp fragment from Lunatic Fringe cDNA was compared with that of a 558 bp fragment of GAPDH used as an internal control. One unaffected and three lesional skin cDNA samples were amplified by PCR looking for the following fragments: (Radical Fringe: 300 bp, Manic Fringe: 622 bp, Notch1: 724 bp, Notch2: 473 bp, Delta1: 301 bp, Jagged1: 509 bp). Sample 1: normal human skin; sample 2: skin from an unaffected region of a psoriatic patient; sample 3: psoriatic skin from the same patient; sample 4: psoriatic skin, treated by phototherapy; sample 5 : infiltrating BCC (see Fig. 3C); sample 6: nodular BCC (see Fig. 3B); sample 7:infiltrating BCC (see Fig. 3D,3E,3F); sample 8: unaffected skin surrounding the excised BCC region (see left side of Fig. 3C).
Moreover in situ experiments on small areas of BCC as well as fringe of wound healing experiments provided an internal control of the studied RNA integrity (Fig 2, solid arrowheads, Fig 3C, arrowhead).
Discussion
At the tissue resolution level, reached with radiolabelled probes, we observe continuous labelling in the basal layer, including putative stem cells and transit amplifying cells. Thus in the epidermis of human adult,Notch and Delta/Jagged ligands, as well as Fringe genes are transcribed in the proliferating area receiving the first differentiation signals. To a lesser extent, they are expressed in the suprabasal layers where differentiation is occurring. No major difference between the expression pattern of Notch1–3, nor those of Delta1/Jagged1 ligands or the Fringe genes are found, except the latter genes are more restricted to the basal layer. The unexpected co-localisation of the mRNA of all Notch partners in the same tissue needs to be reconciled with regards to previous data.
In human skin, Lowell _et al._[6] found that Delta1 mRNA and protein were restricted to the basal epidermal layer while immuno-reactivity against Notch1 protein was patchy in the basal layer and fairly strong in the suprabasal layer. A possible way to accomodate both data regarding Notch1 mRNA (our results) and Notch1 protein (Lowell et al. results) would be to look for possible post-transcriptional and/or post-translational control of protein levels leading to Notch1 protein accumulation in the differentiating keratinocyte. In support of this explanation, we found that Notch1 is still expressed by keratinocytes leaving the basal layer for the suprabasal layer.
In mouse embryo epidermis, each of the three homologous Fringe genes (Lunatic, Radical and Manic) present a specific pattern in the basal and suprabasal layers [2], as do Notch1 and Jagged2 in rat embryonic skin [34]. In the embryonic mouse upper lip, Powell _et al._[5] found that Notch1, Jagged1 and Jagged2 have different expression pattern, which become overlapping in the neonate, as the hair vibrissae follicle bud is developping.
In the adult mouse, these genes are only transcribed at low levels in interfollicular skin, detectable only by PCR [2], and their spatial pattern has not yet been determined. However, in cycling hair follicles of adult mice, transcripts can be weakly detected by in situ hybridisation and display partial overlapping patterns [3], which is reminiscent of our present result in human.
The discrepancy between co-localisation of Notch-related transcripts in adult skin and specific pattern in most other systems may only be apparent, since at a cellular level, ligand and receptor gene expression may be cell-exclusive in the basal layer. Some cells, or clusters of cells, could express the ligands, and neighbouring cells that express only the Notch receptor may behave as responding cells. This was recently demonstrated by Lowell et al. [6] for Delta 1 - Notch 1 in keratinocyte cultures and neonatal foreskin. This creates a boundary between stem cells expressing Delta 1 and cells committed to differentiation, which express Notch 1. We performed cold labelling of probes to assess to cellular level in adult skin samples. Sensitivity of this method was insufficient under our conditions to get conclusive pictures, due to the low level of expression of Notch related genes (data not shown). Alternatively the co-expression of transcripts of the same group (ligand, receptor or modulator) in the same tissue and/or cells may explain the apparent paradox that neither _Notch_-related gene-targeted inactivation experiments in mice [35] nor Notch-related human syndromes [23] produce any skin phenotype. This co-expression of paralogous genes (at least the 3 Notch or the 2 ligands or the 3 Fringe), would lead to their functional redundancy in skin so that the disruption of one of these genes may be substituted by a paralogous one.
When keratinocytes enter into a pathological proliferation status (BCC, psoriasis), they neither transcribe ligands nor receptors and the differentiating signal is absent. It should be noted that the integrity of the mRNAs was checked by RT-PCR of several different samples extracts. A strong positive signal was obtained with GAPDH housekeeping genes, whereas the Lunatic Fringe 309 bp fragment was not amplified in psoriatic and BCC skin compared with controls (Fig. 5). This excludes the possibility that loss of expression of _Notch_-related genes in diseased tissue samples is due to loss of RNA integrity. This observation is strengthened by the results obtained for the expression at the excision margin (Fig. 3C) and at the localised region of basal layer (Fig. 3A,3D,3E) which provide a control. After phototherapy of psoriatic skin, expression of _Notch_-related genes is restored and partial differentiation of keratinocytes occurred. During healing, a rapid but temporary proliferation status of keratinocytes is required to cover the newly constituted granulocytic layer, and they no longer express ligands, nor receptor transcripts. Transcription is then progressively activated when the epidermis starts to become stratified and differentiated.
We show by observation of skin pathologies (BCC, psoriasis), course of treatment (phototherapy) and by using in vivo experimentation (wound healing), that Delta/Notch signalling is decreased in uncontrolled proliferating conditions and conversely, it is triggered as soon as cells are committed to a differentiation programme. We have also shown previously that in anagen vibrissa follicle of adult mice where different cell fates are to be determined for keratinocytes, _Notch_-1, Lunatic and Manic Fringe genes are expressed in specific layers of the outer root sheath. In the catagen phase, when ill-defined and largely undifferentiated keratinocytes fill the follicle, there is no expression of Lunatic and Manic Fringe genes, and _Notch_-1 has a diffuse expression pattern. Hence, in vivo observations, which are at tissue resolution, corroborate in vitro results showing that in the epidermis, Notch signalling promotes differentiation of receiving cells [6]. Nevertheless, compared with many other situations, this link between Notch and differentiation appears paradoxal, i.e. mutations in _Notch 1, 2, 4_[13, 14, 17] or in _Jagged1_[36, 37] genes inducing constitutive Notch activation mediate proliferation of undifferentiated progenitor blood cells or mammary tumourigenesis. Therefore, this pathway may have the opposite effect depending on the cell type concerned and the tissue environment.
In fact, Notch signalling controls cell choices in both invertebrates and vertebrates [7] by inhibiting certain differentiation pathways, thereby permitting cells to either differentiate to an alternative pathway or to self-renew. Feed-back in the Notch pathway amplifies weak stochastic bias between adjacent cells so that even a subtle variation in the amount of signal would generate drastically distinct cell fate. In the epidermal basal layer, Notch receptors are present in transit amplifying cells [6] and the cell-cell signalling system promotes both expansion and differentiation of these cells. Our results are consistent with the possibility that contact-dependent Notch-ligand interactions among neighbouring basal layer cells mutually control their proliferation and initiate their differentiation. In other words, low Notch activity or even none may be permissive for over-growth of poorly differentiated cells, whereas high Notch activity would be expected to control growth and undergo a change of cells fate. Similarly, recent studies have shown that members of the Notch signalling pathway affect neurite outgrowth and neuronal differentiation [12]. Notch/Delta is also critical for the decision between the endocrine and exocrine fates in developing pancreas [10]. It has been shown that despite their endodermal origin, pancreatic cells share several molecular characteristics with neurones. Our results are also consistent with the observation of reactivation of Notch and Delta 1 during cell differentiation consecutive to injury of mouse dental pulp [38].
The broad down-regulation of _Notch_-related genes expression in diseased skin raised the question as to whether this phenomenon occurs with other mRNAs. A broad-scale evaluation of gene expression has been reported in scar tissue compared with normal skin using a cDNA microarray of 4 000 genes without indication whether Notch-related genes were included [39]. Data indicate that 142 genes were over expressed compared with uninjured skin, while 50 were downregulated. Another experimental model comparing psoriatic-involved skin and normal control skin by a differential display strategy showed around 60 cDNA species to be specifically displayed in psoriasis [40], but downregulated genes were not the focus of this approach. To our knowledge, no extensive studies have been published concerning transcriptome behaviour in the case of BCCs compared with normal control skin or paired adjacent skin. This is probably because BCC have been shown to be associated with mutations that activate the proto-oncogene Smoothened (SMO) or that inactivate the tumour suppressor Patched (PTCH), both involved in the Hedgehog (Hh) pathway. Therefore, BCC is known to be initiated by constitutive activation of _Hedgehog_-responsive genes [27], excluding Notch dysfunction as the initial event. Therefore, the subsequent involvement of Notch in this disorder remains to be investigated. Numerous proto-oncogenes, cytokines and signalling molecule mRNA expression have been recently evaluated by hybridisation [40–44], Together, these studies show that the down-regulation of Notch/Delta genes cannot account for a global down-regulation of most mRNA synthesis following skin injury or disease. However, as yet only little is known concerning the Notch/Delta pathway in skin disorders. Interestingly, Manic Fringe maps in the region of two cancer-associated loci and Radical Fringe maps close to a familial psoriasis susceptibility locus [45].
Conclusions
We show here that Delta, Notch and Fringe signalling molecules are up-regulated in the basal and differentiating layers of adult human epidermis, whereas they are under-expressed in disorganised proliferating epidermis. We postulate that the specific differentiation programme that keratinocytes undergo in skin is under their control. We suggest that alteration of the Notch pathway may somehow play a role in disrupting the keratinocyte differentiation programme, leading to unorganised suprabasal epidermal strata and/or dermal invasion. In the cases studied here, it remains to be definitively established whether their transcription is a cause rather than a consequence of commitment to epidermal differentiation.
References
- Del Amo FF, Smith DE, Swiatek PJ, Gendron-Maguire M, Greenspan RJ, McMahon AP, Gridley T: Expression pattern of Motch, a mouse homolog of Drosophila Notch, suggests an important role in early postimplantation mouse development. Development 1992, 115: 737–744.
CAS PubMed Google Scholar - Thélu J, Viallet JP, Dhouailly D: Differential expression pattern of the three Fringe genes is associated with epidermal differentiation. J Invest Dermatol 1998, 111: 903–906. 10.1046/j.1523-1747.1998.00372.x
Article PubMed Google Scholar - Favier B, Flignaux I, Thélu J, Viallet JP, Demarchez M, Jahoda CAB, Dhouailly D: The notch system and the differentiation of vibrissa hair follicule: receptors, ligands and fringe modulators. Developmental Dynamics 2000, 218: 426–437. 10.1002/1097-0177(200007)218:3<426::AID-DVDY1004>3.3.CO;2-W
Article CAS PubMed Google Scholar - Kopan R, Weintraub H: Mouse notch: expression in hair follicles correlates with cell fate determination. J Cell Biol 1993, 121: 631–641. 10.1083/jcb.121.3.631
Article CAS PubMed Google Scholar - Powell BC, Passmore EA, Nesci A, Dunn SM: The Notch signalling pathway in hair growth. Mech Dev 1998, 78: 189–192. 10.1016/S0925-4773(98)00177-4
Article CAS PubMed Google Scholar - Lowell S, Le Roux I, Dunne J, Watt FM: Stimulation of human epidermal differentiation by Delta-Notch signalling at the boundaries of stem-cell clusters. Current Biology 2000, 10: 491–500. 10.1016/S0960-9822(00)00451-6
Article CAS PubMed Google Scholar - Artavanis-Tsakonas S, Rand MD, Lake RJ: Notch signalling: cell fate control and signal integration in development. Science 1999, 284: 770–776. 10.1126/science.284.5415.770
Article CAS PubMed Google Scholar - Egan SE, St-Pierre B, Leow CC: Notch receptors, partners and regulators: from conserved domains to powerful functions. Curr Top Microbiol Immunol 1998, 228: 273–324.
CAS PubMed Google Scholar - Panin VM, Irvine KD: Modulators of Notch signaling. Cell Dev Biol 1998, 9: 609–617. 10.1006/scdb.1998.0263
Article CAS Google Scholar - Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T, Hrabe de Angelis M, Lendahl U, Edlund H: Notch signalling controls pancreatic cell differentiation. Nature 1999, 400: 877–881. 10.1038/23716
Article CAS PubMed Google Scholar - Milner LA, Bigas A: Notch as a mediator of cell fate determination in hematopoiesis: evidence and speculation. Blood 1999, 93: 2431–2448.
CAS PubMed Google Scholar - Sestan N, Artavanis-Tsakonas S, Rakic P: Contact-dependent inhibition of cortical neurite growth mediated by notch signaling. Science 1999, 286: 741–746. 10.1126/science.286.5440.741
Article CAS PubMed Google Scholar - Capobianco AJ, Zagouras P, Blaumueller CM, Artavanis-Tsakonas S, Bishop JM: Neoplastic transformation by truncated alleles of human NOTCH1/TAN1 and NOTCH2. Mol Cell Biol 1997, 17: 6265–6273.
Article CAS PubMed PubMed Central Google Scholar - Milner LA, Bigas A, Kopan R, Brashem-Stein C, Bernstein ID, Martin DI: Inhibition of granulocytic differentiation by mNotch1. Proc Natl Acad Sci U S A 1996, 93: 13014–13019. 10.1073/pnas.93.23.13014
Article CAS PubMed PubMed Central Google Scholar - Bray S: A Notch affair. Cell 1998, 93: 499–503. 10.1016/S0092-8674(00)81180-0
Article CAS PubMed Google Scholar - Joutel A, Corpechot C, Ducros A, Vahedi K, Chabriat H, Mouton P, Alamowitch S, Domenga V, Cecillion M, Marechal E, Maciazek J, Vayssiere C, Cruaud C, Cabanis EA, Ruchoux MM, Weissenbach J, Bach JF, Bousser MG, Tournier-Lasserve E: Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature 1996, 383: 707–710. 10.1038/383707a0
Article CAS PubMed Google Scholar - Gallahan D, Callahan R: The mouse mammary tumor associated gene INT3 is a unique member of the NOTCH gene family (NOTCH4). Oncogene 1997, 14: 1883–1890. 10.1038/sj/onc/1201035
Article CAS PubMed Google Scholar - Smith GH, Gallahan D, Diella F, Jhappan C, Merlino G, Callahan R: Constitutive expression of a truncated INT3 gene in mouse mammary epithelium impairs differentiation and functional development. Cell Growth Differ 1995, 6: 563–577.
CAS PubMed Google Scholar - Krantz ID, Colliton RP, Genin A, Rand EB, Li L, Piccoli DA, Spinner NB: Spectrum and frequency of jagged1 (JAG1) mutations in Alagille syndrome patients and their families. Am J Hum Genet 1998, 62: 1361–1369. 10.1086/301875
Article CAS PubMed PubMed Central Google Scholar - Oda T, Elkahloun AG, Pike BL, Okajima K, Krantz ID, Genin A, Piccoli DA, Meltzer PS, Spinner NB, Collins FS, Chandrasekharappa SC: Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat Genet 1997, 16: 235–242.
Article CAS PubMed Google Scholar - Sun X, Artavanis-Tsakonas S: The intracellular deletions of Delta and Serrate define dominant negative forms of the Drosophila Notch ligands. Development 1996, 122: 2465–2474.
CAS PubMed Google Scholar - May WA, Arvand A, Thompson AD, Braun BS, Wright M, Denny CT: EWS/FLI1-induced manic fringe renders NIH 3T3 cells tumorigenic. Nat Genet 1997, 17: 495–497.
Article CAS PubMed Google Scholar - Joutel A, Tournier-Lasserve E: Notch signalling pathway and human diseases. Semin Cell Dev Biol 1998, 9: 619–625. 10.1006/scdb.1998.0261
Article CAS PubMed Google Scholar - Lang PG, Maize JC: Basal cell carcinoma. Philadelphia: Saunders, W.B. 1991.
Google Scholar - Miller SJ: Biology of basal cell carcinoma (Part I). J Am Acad Dermatol 1991, 24: 1–13.
Article CAS PubMed Google Scholar - Miller SJ: Biology of basal cell carcinoma (Part II). J Am Acad Dermatol 1991, 24: 161–175.
Article CAS PubMed Google Scholar - Wicking C, Smyth I, Bale A: The hedgehog signalling pathway in tumorigenesis and development. Oncogene 1999, 18: 7844–7851. 10.1038/sj/onc/1203282
Article CAS PubMed Google Scholar - Chopra A, Maitra B, Korman NJ: Decreased mRNA expression of several basement membrane components in basal cell carcinoma. J Invest Dermatol 1998, 110: 52–56. 10.1046/j.1523-1747.1998.00089.x
Article CAS PubMed Google Scholar - Wright NA, Camplejohn RS: Cell proliferation. Edinburg: Churchill Livingstone 1985.
Google Scholar - McKay IA, Leigh IM: Altered keratinocyte growth and differentiation in psoriasis. Clin Dermatol 1995, 13: 105–114. 10.1016/0738-081X(95)93817-8
Article CAS PubMed Google Scholar - Mansbridge JN, Knapp AM: Changes in keratinocyte maturation during wound healing. J Invest Dermatol 1987, 89: 253–263.
Article CAS PubMed Google Scholar - Viallet JP, Dhouailly D: Retinoic acid and mouse skin morphogenesis. I. Expression pattern of retinoic acid receptor genes during hair vibrissa follicle, plantar, and nasal gland development. J Invest Dermatol 1994, 103: 116–121.
Article CAS PubMed Google Scholar - Demarchez M, Hartmann DJ, Herbage D, Ville G, Pruniéras M: Wound healing of human skin transplanted onto the nude mouse : an immunohistological study and ultrastructural study of the epidermal basement membrane zone reconstruction and connective tissue reorganization. Dev Biol 1987, 121: 119–129.
Article CAS PubMed Google Scholar - Shawber C, Boulter J, Lindsell CE, Weinmaster G: Jagged2: a serrate-like gene expressed during rat embryogenesis. Dev Biol 1996, 180: 370–376. 10.1006/dbio.1996.0310
Article CAS PubMed Google Scholar - Huppert SS, Le A, Schroeter EH, Mumm JS, Saxena MT, Milner NA, Kopan R: Embryonic lethality in mice homozygous for a processing-deficient allele of Notch1. Nature 2000, 405: 966–970. 10.1038/35016111
Article CAS PubMed Google Scholar - Moore KA, Pytowski B, Witte L, Hicklin D, Lemischka IR: Hematopoietic activity of a stromal cell transmembrane protein containing epidermal growth factor-like repeat motifs. Proc Natl Acad Sci U S A 1997, 94: 4011–4016. 10.1073/pnas.94.8.4011
Article CAS PubMed PubMed Central Google Scholar - Varnum-Finney B, Purton LE, Yu M, Brashem-Stein C, Flowers D, Staats S, Moore KA, Le Roux I, Mann R, Gray G, Artavanis-Tsakonas S, Bernstein ID: The Notch ligand, Jagged-1, influences the development of primitive hematopoietic precursor cells. Blood 1998, 91: 4084–4091.
CAS PubMed Google Scholar - Mitsiadis TA, Fried K, Goridis C: Reactivation of Delta-Notch signaling after injury: complementary expression patterns of ligand and receptor in dental pulp. Exp Cell Res 1999, 246: 312–318. 10.1006/excr.1998.4285
Article CAS PubMed Google Scholar - Tsou R, Cole JK, Nathens AB, Isik FF, Heimbach DM, Engrav LH, Gibrans NS: Analysis of hypertrophic and normal scar gene expression with cDNA microarrays. J Burn Care Rehabil 2000, 21: 541–550.
Article CAS PubMed Google Scholar - Bockelmann R, Neugebauer P, Paseban ND, Huttemann M, Gollnick H, Bonnekoh B: Suprabasal overexpression of the hsRPB7 gene in a psoriatic epidermis as identified by a reverse transcriptase-polymerase chain reaction differential display model comparing psoriasis plaque tissue with peritonsillar mucosa. Am J Pathol 2001, 158: 367–372.
Article CAS PubMed PubMed Central Google Scholar - Duvic M, Helekar B, Schulz C, Cho M, DiSepio D, Hager C, DiMao D, Hazarika P, Jackson B, Breuer-McHam J, Clayman G, Lippman SM, Chandraratna RA, Robinson NA, Deutcher A, Eckert RL, Nagpal S: Expression of a retinoid-inducible tumor suppressor, Tazarotene-inducible gene-3, is decreased in psoriasis and skin cancer. Clin Cancer Res 2000, 6: 3249–3259.
CAS PubMed Google Scholar - Beer HD, Gassmann MG, Munz B, Steiling H, Engelhardt F, Bleuel K, Werner S: Expression and function of keratinocyte growth factor and activin in skin morphogenesis and cutaneous wound repair. J Investig Dermatol Symp Proc 2000, 5: 34–39. 10.1046/j.1087-0024.2000.00009.x
Article CAS PubMed Google Scholar - Ramdial PK, Madaree A, Reddy R, Chetty R: bcl-2 protein expression in aggressive and non-aggressive basal cell carcinomas. J Cutan Pathol 2000, 27: 283–291. 10.1034/j.1600-0560.2000.027006283.x
Article CAS PubMed Google Scholar - Asumalahti K, Laitinen T, Itkonen-vatjus R, Lokki ML, Suomela S, Snellman E, Saarialho-Kere U, Kere J: A candidate gene for psoriasis near HLA-C, HCR(Pg8), is highly polymorphic with a disease-associated susceptibility allele. Human Mol Genet 2000, 9: 1533–1542. 10.1093/hmg/9.10.1533
Article CAS Google Scholar - Moran JL, Johnston SH, Rauskolb C, Bhalerao J, Bowcock AM, Vogt TF: Genomic structure, mapping, and expression analysis of the mammalian Lunatic, Manic, and Radical fringe genes. Mamm Genome 1999, 10: 535–541. 10.1007/s003359901039
Article CAS PubMed Google Scholar
Pre-publication history
- The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-5945/2/7/prepub