Different evolutionary patterns between young duplicate genes in the human genome (original) (raw)
Background
Since Ohno's work [1] gene duplication is widely believed to be the major source of genetic novelties. However, how the two duplicate genes evolve after the duplication event and what the major factors are that determine the fate of duplicate genes remain poorly understood and are currently under intense research.
Lynch and Conery [2] conducted a study of several eukaryotic genomes and concluded that duplicate genes often experience relaxed functional constraints and accumulate mutations at an accelerated rate. However, since their study used within-genome data without an outgroup, it could not reveal differences in evolutionary patterns between two duplicates. Hughes and Hughes [3] used human genes as outgroups to examine 17 pairs of duplicated frog genes, most of which have been duplicated recently. They found that the two duplicate copies of a gene had evolved at approximately the same rate. However, human genes are only distantly related to frog genes and may not be suitable outgroups for young duplicate frog genes. In contrast, Robinson-Rechavi and Laudet [4] and Van de Peer et al. [5], who used human or mammalian genes as outgroups to zebrafish genes, found evidence of unequal evolutionary rates between duplicate genes in zebrafish, although the percentage of pairs with such a pattern differed between the two studies. Furthermore, in a study of young duplicate genes in humans and rodents, Kondrashov et al. [6] found only two out of 49 duplicate genes showed different rates of evolution. Thus, the issue remains to be resolved.
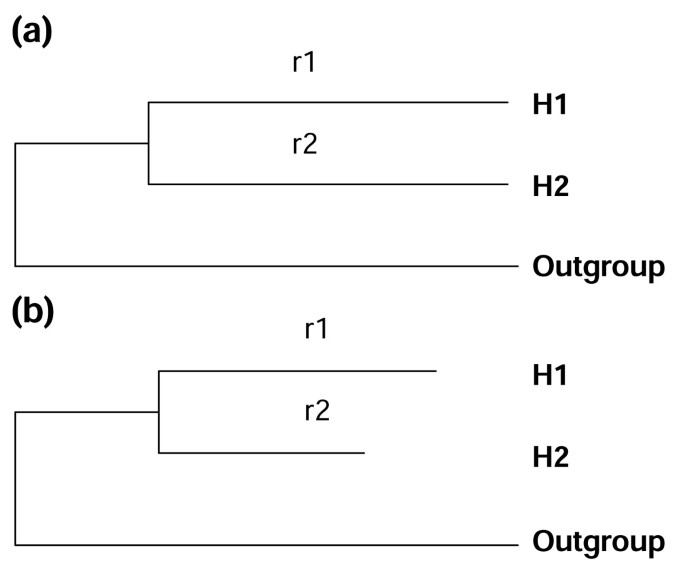
We are interested in the questions of whether duplicated genes in general undergo different evolutionary patterns and what the possible causes for this could be. To address these questions, young human duplicate genes, which are defined as duplicate genes with Ks < 0.3, are excellent materials for several reasons. Firstly, in young duplicates the Ks (the number of synonymous substitutions per synonymous site) and Ka (the number of nonsynonymous substitutions per nonsynonymous site) values are small and can be estimated more accurately than in older duplicates. Secondly, human genes usually have no strong codon usage bias, so the Ks values are not strongly distorted by this effect. Thirdly, the mouse genome provides a suitable outgroup. With the use of outgroup sequences, the maximum likelihood method [7] can be applied, which allows the comparison of various rate models [8]. Different models for amino-acid sequence evolution, with outgroups incorporated, can be compared to judge whether the amino-acid substitution rates are the same in two duplicate copies (Figure 1). Similarly, the models for coding sequences can also be compared to judge if the Ka/Ks ratios are the same in the two copies after a gene duplication. Traditionally, Ka/Ks is taken as an index for the strength of functional constraints. Different Ka/Ks ratios usually suggest different functional constraints on two duplicate copies.
Figure 1
Two models of protein sequence evolution. By comparing the likelihood values of these two models, one can judge whether r1 = r2 (r1 and r2 are the amino-acid substitution rates on branches H1 and H2, respectively). (a) The model assumes r1 = r2. (b) The model allows r1 and r2 to be different. H1 and H2: two human duplicate genes. Outgroup: the mouse ortholog.
Another way to examine if two duplicate copies have experienced different functional constraints is to see whether the distribution of substitutions along their sequences are the same. If a duplicate copy is free of functional constraints, then amino-acid substitutions should occur evenly across the sequence. On the other hand, if a duplicate copy is still under considerable functional constraints, then functionally important regions should be subject to stronger constraints than functionally less important regions, and will accumulate fewer substitutions, thus yielding an uneven substitution pattern. Tang and Lewontin [9] described a statistical method to give a quantitative measure for distinguishing between even and uneven substitution patterns. The rationale of this method is that if substitutions occur evenly across a sequence, a cluster of short spaces (lengths between two consecutive substitutions) should not be extremely long. To test the significance, the longest stretch where every space is short is compared to a simulated distribution generated under the hypothesis of even substitution pattern. We combined Tang and Lewontin's [9] method and the maximum likelihood method of locating substitutions [7] to test the evenness of the substitution patterns of two duplicate genes.
Results and discussion
Amino-acid substitution rates
We examined first whether the amino-acid substitution rates in two duplicate copies are the same. We found that among the 250 pairs of young human duplicates studied, 145 pairs showed significant evidence (at the 5% level) that one copy had evolved faster than the other at the amino-acid level. Among them, 130 pairs had significantly different rates at the 1% significance level.
Hughes and Hughes [3] found similar evolutionary rates in the 17 frog duplicates they studied, probably because the human outgroup they used was too distant to make the statistical test powerful. Using fairly closely related outgroups, our results suggest that the majority of young human duplicates evolve at different rates. Our results are consistent with those of Van de Peer et al. [5], but the number of gene pairs with significantly unequal rates is much higher than Robinson-Rechavi and Laudet [4] (four out of 19) and Kondrashov et al. [6] (two out of 49) found.
Ka/Ksratio
To discover if the functional constraints were the same on two duplicate genes, we examined the Ka/Ks ratio on each branch leading to the two copies. Among the 250 pairs, 65 pairs showed significantly different Ka/Ks ratios at the 5% level and 31 pairs showed a significant difference at the 1% level. As mentioned earlier, the Ka/Ks ratio is an important index of functional constraints. The smaller the Ka/Ks ratio is, the stronger the functional constraints are. Our result suggests that after gene duplication, a substantial proportion (65/250 = 26%) of the duplicate pairs have experienced different functional constraints.
Among the 65 pairs that have different Ka/Ks ratios, 54 pairs also differ between the two copies in their amino-acid substitution rates. Among the 185 pairs that showed no significant difference in Ka/Ks ratios, less than 50% showed significantly different amino-acid substitution rates. A 2 × 2 chi-square test (Table 1, χ2 = 22.675, df = 1, p < 0.001) reveals a significant correlation between different Ka/Ks ratios and different amino-acid substitution rates. Therefore, duplicate pairs with different Ka/Ks ratios tend to evolve at different rates, suggesting that different functional constraints might be largely responsible for the unequal evolutionary rates, although, as mentioned below, some duplicate genes have apparently undergone positive selection.
Table 1 Substitution rates versus Ka/Ks ratios in duplicate genes
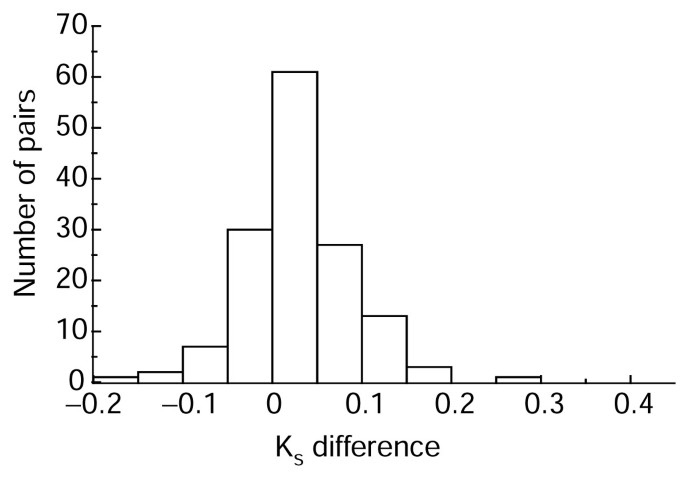
One reason why we could not detect as many pairs with different Ka/Ks ratios as pairs with different amino-acid substitution rates could be, in part, because fast-evolving sequences tend to have a higher Ks than slow-evolving ones. To see whether this was true, we calculated the Ks difference between the two copies of each pair - the Ks of the fast-evolving copy minus the Ks of the slow-evolving copy. Figure 2 shows that most of the pairs have a positive Ks difference, which means that in most of the pairs the fast-evolving copy has a higher Ks than the slow-evolving copy.
Figure 2
Distribution of Ks differences between duplicate genes for gene pairs with different amino-acid substitution rates. The x axis is the Ks difference between duplicate genes (the Ks of the fast-evolving copy minus the Ks of the slow-evolving one). The y axis is the number of gene pairs within a Ks bin. This figure shows that most pairs have a positive Ks difference, which suggests that the fast-evolving copy usually has a higher Ks than the slow-evolving copy.
Two duplicate copies may differ significantly in the number of amino-acid substitutions, which reflects a significant difference between two Ka values at the nucleotide level. However, the two Ks values are also different (usually the copy with a higher Ka also has a higher Ks) which reduces the chance for the Ka/Ks ratios of the two copies to be significantly different. This weak correlation between Ka and Ks is consistent with several previous studies [10–13] and may be largely explained by the fact that silent sites in some genes are also under purifying selection (that is, codon usage bias) [14]. So, although we found nearly 60% of pairs with different amino-acid substitution rates, we found far fewer pairs having different Ka/Ks ratios.
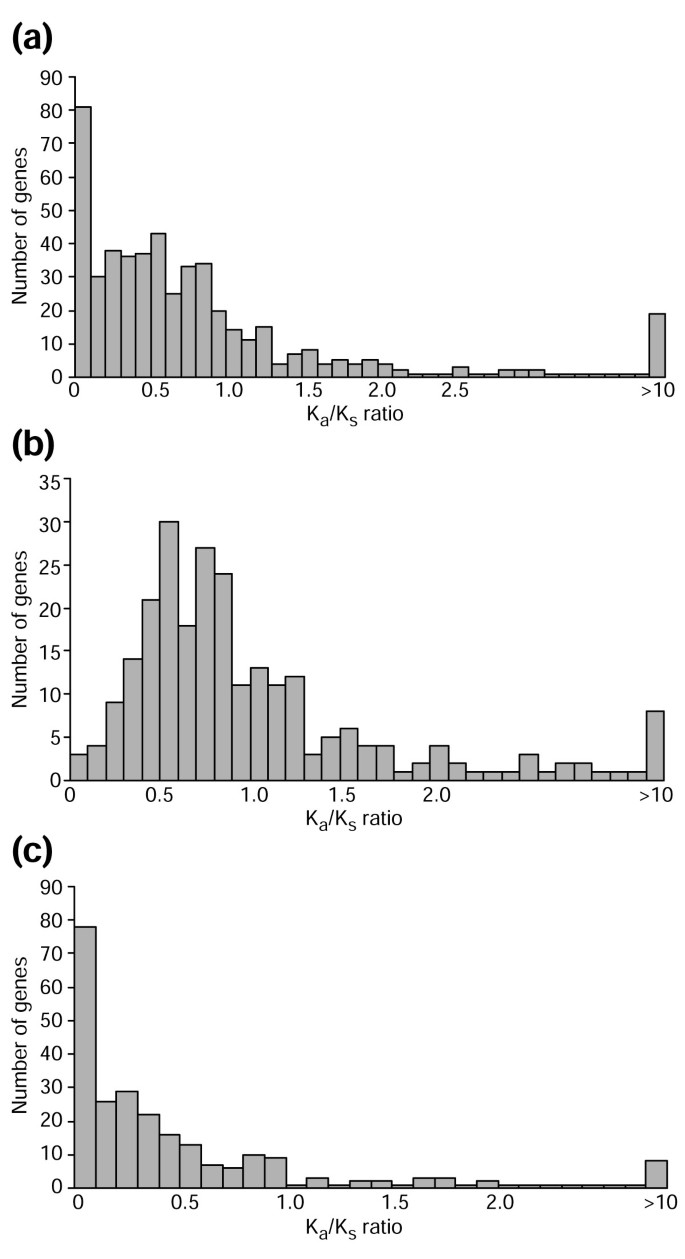
We also looked for evidence of positive selection. Figure 3a shows that most of the genes have a Ka/Ks ratio of less than one, although there are still 113 genes with a Ka/Ks ratio greater than one. Ka/Ks > 1 suggests positive selection but evidence for positive selection requires the ratio to be significantly greater than one. In the genes with Ka/Ks > 1, many results are just slightly greater than one and only seven genes are found to have the Ka/Ks ratio significantly greater than one. However, this does not imply that only seven pairs of duplicate genes were subject to positive selection because, in many cases, the number of substitutions between two young duplicates may be too small for the test to be statistically significant, even if some of the substitutions have occurred by positive selection.
Figure 3
The Ka/Ks ratio distribution of young human duplicates. The x axis is the Ka/Ks ratio on the branch leading to one human duplicated gene. The y axis is the number of genes within a Ka/Ks bin. (a) All genes from the 250 pairs, a total of 500 sequences. (b) The fast-evolving duplicate copies of 250 pairs, a total of 250 sequences. (c) The slow-evolving duplicate copies of 250 pairs, a total of 250 sequences.
Most of the fast-evolving duplicate copies have higher Ka/Ks ratios (Figure 3b) than slow-evolving duplicate copies (Figure 3c). This supports the view that after gene duplication, one duplicate copy may have undergone purifying selection, while the functional constraints on the other copy may have been relaxed to some extent.
Different substitution patterns
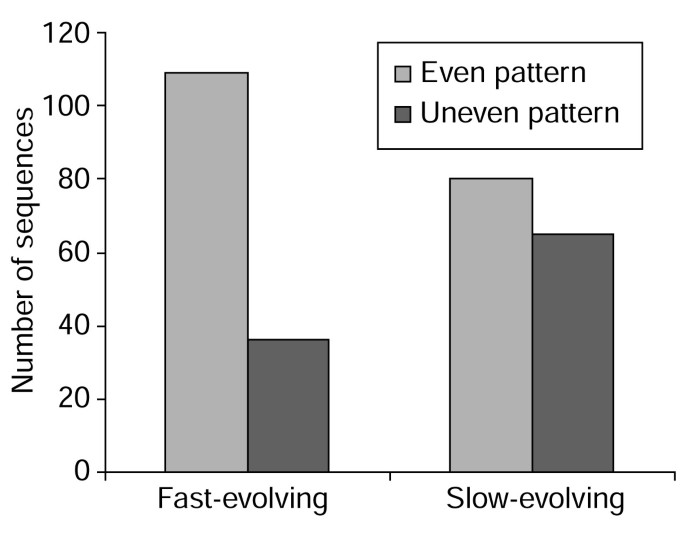
Among the 145 fast-evolving human young duplicates, 109 have an even amino-acid substitution pattern across the sequence between the human and mouse orthologs. In other words, these 109 sequences show no large highly-conserved regions. On the other hand, 65 of the 145 slow-evolving copies show evidence of an uneven substitution pattern between human and mouse orthologs, which suggests that they have some slow-evolving regions and some fast-evolving regions at the protein level.
In order to infer the position of each amino-acid substitution in the sequence, we inferred the ancestral sequences by using PAML (Phylogenetic Analysis by Maximum Likelihood) [7] standard settings, which assume constant rates across sites. It is possible, therefore, that our estimated substitutions may be more evenly distributed than they actually are. However, because we are comparing the percentage of sequences with even patterns in fast-evolving copies to those in slow-evolving copies, this potential bias should be on both sides of the comparison and should not change our conclusion.
Figure 4 with a chi-square test (χ2 = 12.78, df = 1, p < 0.01) shows that fast-evolving duplicates have a significantly higher proportion of sequences with an even substitution pattern. This finding suggests that most of the fast-evolving copies have more relaxed functional constraints than slow-evolving copies and tend to accumulate substitutions evenly across the sequence. The suggestion of relaxed functional constraints for young duplicates is consistent with the observation of Lynch and Conery [2]. Of course, we cannot exclude the possibility that some of the amino-acid substitutions in fast-evolving copies might have been due to positive Darwinian selection.
Figure 4
Comparison between fast-evolving copies and slow-evolving copies. The figure shows that fast-evolving copies have more cases with substitutions distributed evenly along the sequence than slow-evolving copies. Fast-evolving: the copy that has evolved faster than the other in each duplicate pair. Slow-evolving: the copy that has evolved slower than the other in each duplicate pair. Even pattern: a sequence that has evenly-distributed substitutions along the sequence. Uneven pattern: a sequence that has unevenly-distributed substitutions.
Our finding is very different from that of Kondrashov et al. [6], who found only two pairs with unequal evolutionary rates out of 49 pairs studied in mammals. Since they also focused on young duplications (0.05 < Ks < 0.5) and the approach they used to identify duplicate genes is similar to ours, this may be due to the different datasets used. Since the neutral pattern found in the fast-evolving copies in our study is to some extent similar to the evolution of pseudogenes, we examined the possibility of the inclusion of many pseudogenes in our sample.
The gene predictions in the Ensembl database [15] we used always produce a translation for each gene and a stringent criterion (near full-length similarity) was used in our grouping method; consequently, our dataset does not include pseudogenes due to premature codons. Since we limited our set of duplicated genes to Ks > 0.05, a pseudogene in our sample would be likely to have lost its function only very recently, otherwise it would have gained one or more premature stop codons since the time of nonfunctionalization. In the Ensembl database we used only those genes ('known' genes) with experimental support and those genes ('novel' genes) with high similarity to known genes in human and other organisms. Genes purely from Genscan predictions were not used in this analysis. These approaches would have effectively reduced the portion of pseudogenes in our dataset. If the functional constraints on a gene are largely relaxed, the evolutionary pattern of this gene may be similar to that of pseudogenes. So it is possible that some of the fast-evolving genes may be on their way to become pseudogenes, although it is still possible that they may evolve new functions. Kondrashov et al. [6] used a cDNA-based dataset and found only a few duplicated pairs with different evolutionary rates, which may have represented those genes that survived well through selection and were still functioning. In other words, the cDNA-based genes which they used are normally expressed, meaning these genes may still be under strong selection pressure. Our dataset might be more appropriate for providing a general scenario of how two duplicate genes evolve after gene duplication.
Conclusion
We used conservative criteria to select young human duplicate pairs and applied a stringent statistical method to test whether two duplicate copies exhibit different evolutionary patterns. Our results suggest that, in most cases, during the early stage of evolution following gene duplication, the two duplicates evolve at different rates, which could affect the fate of the two copies. Different functional constraints on the two copies may have been largely responsible for the different rates. One copy may have relaxed functional constraints, while the other could still be under strong constraints. The stringent statistical tests used in this study might have underestimated the proportion of pairs with this pattern, but this could only strengthen our argument.
Materials and methods
Processing data and selecting independent young human duplicate genes
Human genes were downloaded from the Ensembl human database [15] version 11.31 (28 February 2003). The original dataset is available from the authors on request. Only known and novel genes were used in this analysis; those sequences containing repetitive elements detected by FASTA (E = 10-5) searching against Repbase [16–[18](/article/10.1186/gb-2003-4-9-r56#ref-CR18 "Repbase. [ http://www.girinst.org/Repbase_Update.html
]")\] were removed from the dataset. If two genes overlapped at a chromosomal position, the gene with the longer protein was retained. The protein sequences selected were grouped into families by the method used by Gu _et al_. \[[19](/article/10.1186/gb-2003-4-9-r56#ref-CR19 "Gu Z, Cavalcanti A, Chen FC, Bouman P, Li WH: Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol Biol Evol. 2002, 19: 256-262.")\]. Within each gene family, the selection of independent duplicate gene pairs proceeded with increasing Ks. That is, within each gene family, we selected the gene pair with the smallest Ks and excluded it from the family and then selected the gene pair with the smallest Ks from among the remaining genes. We repeated this until no gene pairs could be selected. This method ensured that a gene in one pair would not appear in another pair. Among the duplicate genes we selected, those pairs with 0.05 < Ks < 0.3 were used in this study. We used Ks < 0.3 as a cutoff to define young human duplicates. With the Ks between human duplicate genes less than 0.3, the duplication would have occurred less than 50 million years ago. Pairs with Ks < 0.05 have too few substitutions to make a statistical test meaningful. For each pair we selected, both copies were used to search the mouse database. Mouse genes were also obtained from the Ensembl database \[[15](/article/10.1186/gb-2003-4-9-r56#ref-CR15 "Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, et al: The Ensembl genome database project. Nucleic Acids Res. 2002, 30: 38-41. 10.1093/nar/30.1.38.")\] version 11.3 (28 February 2003) and were cleaned using the same procedure that was used to clean the human database. The pairs in which the two copies had the same best hits and a human-mouse Ks < 1 were kept for this study. We chose Ks < 1 as a cutoff point because a distant outgroup makes it harder to detect rate differences. A set of 250 young human duplicate pairs and their mouse orthologs were thus retained.Statistical methods to compare evolutionary patterns between two copies
To calculate if the evolutionary rates and the Ka/Ks ratio are the same between the two duplicate copies, the likelihood-ratio test [8] was applied to each pair selected. To test the hypothesis of equal evolutionary rates between the two duplicate copies at the amino-acid level, a two-rate model and a free-rate model were compared. The two models differ in that the two-rate model assumes the same evolutionary rate on the two branches leading to the two duplicates but allows the rate on the outgroup branch to be different, while the free-rate model does not impose any equal rates among branches. The codeml program (set seqtype = 2 for amino-acid sequences) in the PAML package was run for each of the two models with all parameters set to default except for the parameter 'model' for amino-acid substitution, which was set to 'Poisson'. We also set this parameter to the 'Jones-Taylor-Thornton model' and the conclusion was basically the same. Two maximum likelihood values for the two models were given, and twice the difference was compared to a chi-square distribution. If significant, the results suggest that the two branches have evolved at unequal rates. To test if the Ka/Ks ratios are different between the coding sequences of the two duplicates, a two-ratio model, which assumes the same Ka/Ks ratio on the branches leading to the two duplicates but an independent Ka/Ks ratio on the branch leading to the outgroup, was compared to the free-ratio model, which assumes an independent Ka/Ks ratio for each branch.
Substitution patterns and differential selection
Using the PAML package [7], the ancestral sequence of each human duplicate gene pair was reconstructed and the position of each substitution was located. Tang and Lewontin's [9] method was then applied to calculate the T statistics of each human sequence. For each human sequence, 100,000 pseudo-sequences were generated under the null hypothesis that the substitutions distribute evenly across the sequences, and the distribution of T statistics of these 100,000 pseudo-sequences was generated. The T statistic of the real sequence was then compared to this distribution. If the T statistic was extremely large or small (the rejection level is 0.05), we considered it to be an even distribution. The program was written in Perl and is available upon request.
Additional data files
The amino-acid alignments (Additional data file 1) and coding sequence alignments (Additional data file 2) are available in PAML format.
References
- Ohno S: Evolution by Gene Duplication. 1970, Evolution by Gene Duplication. Berlin: Springer-Verlag
Book Google Scholar - Lynch M, Conery JS: The evolutionary fate and consequences of duplicate genes. Science. 2000, 290: 1151-1155. 10.1126/science.290.5494.1151.
Article PubMed CAS Google Scholar - Hughes MK, Hughes AL: Evolution of duplicate genes in a tetraploid animal, Xenopus laevis. Mol Biol Evol. 1993, 10: 1360-1369.
PubMed CAS Google Scholar - Robinson-Rechavi M, Laudet V: Evolutionary rates of duplicate genes in fish and mammals. Mol Biol Evol. 2001, 18: 681-683.
Article PubMed CAS Google Scholar - Van de Peer Y, Taylor JS, Braasch I, Meyer A: The ghost of selection past: rates of evolution and functional divergence of anciently duplicated genes. J Mol Evol. 2001, 53: 436-446. 10.1007/s002390010233.
Article PubMed CAS Google Scholar - Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV: Selection in the evolution of gene duplications. Genome Biol. 2002, 3: research0008.1-0008.9. 10.1186/gb-2002-3-2-research0008.
Article Google Scholar - Yang Z: PAML: a program package for phylogenetic analysis by maximum likelihood. CABIOS. 1997, 13: 555-556.
PubMed CAS Google Scholar - Yang Z, Nielsen R: Synonymous and nonsynonymous rate variation in nuclear genes of mammals. J Mol Evol. 1998, 46: 409-418.
Article PubMed CAS Google Scholar - Tang H, Lewontin RC: Locating regions of differential variability in DNA and protein sequences. Genetics. 1999, 153: 485-495.
PubMed CAS PubMed Central Google Scholar - Graur D: Amino acid composition and the evolutionary rates of protein-coding genes. J Mol Evol. 1985, 22: 53-62.
Article PubMed CAS Google Scholar - Wolfe KH, Sharp PM: Mammalian gene evolution: nucleotide sequence divergence between mouse and rat. J Mol Evol. 1993, 37: 441-456.
Article PubMed CAS Google Scholar - Mouchiroud D, Gautier C, Bernardi G: Frequencies of synonymous substitutions in mammals are gene-specific and correlated with frequencies of nonsynonymous substitutions. J Mol Evol. 1995, 40: 107-113.
Article PubMed CAS Google Scholar - Ohta T, Ina Y: Variation in synonymous substitution rates among mammalian genes and the correlation between synonymous and nonsynonymous divergence. J Mol Evol. 1995, 41: 717-720.
PubMed CAS Google Scholar - Hellmann I, Zollner S, Enard W, Ebersberger I, Nickel B, Paabo S: Selection on human genes as revealed by comparisons to chimpanzee cDNA. Genome Res. 2003, 13: 831-7. 10.1101/gr.944903.
Article PubMed CAS PubMed Central Google Scholar - Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, et al: The Ensembl genome database project. Nucleic Acids Res. 2002, 30: 38-41. 10.1093/nar/30.1.38.
Article PubMed CAS PubMed Central Google Scholar - Jurka J: Repbase Update: a database and an electronic journal of repetitive elements. Trends Genet. 2000, 16: 418-420. 10.1016/S0168-9525(00)02093-X.
Article PubMed CAS Google Scholar - Jurka J: Repeats in genomic DNA: mining and meaning. Curr Opin Struct Biol. 1998, 8: 333-337. 10.1016/S0959-440X(98)80067-5.
Article PubMed CAS Google Scholar - Repbase. [http://www.girinst.org/Repbase_Update.html]
- Gu Z, Cavalcanti A, Chen FC, Bouman P, Li WH: Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol Biol Evol. 2002, 19: 256-262.
Article PubMed CAS Google Scholar