Accessibility of transcriptionally inactive genes is specifically reduced at homeoprotein-DNA binding sites in Drosophila (original) (raw)
Abstract
We showed previously that homeoproteins bind to multiple DNA sites throughout the length of most genes in Drosophila embryos. Based on a comparison of in vivo and in vitro DNA binding specificities, we suggested that homeoprotein binding sites on actively transcribed genes are largely accessible, whereas the binding of homeoproteins to inactive and poorly transcribed genes may be significantly inhibited at most sites, perhaps by chromatin structure. To test this model, we have measured the accessibility of restriction enzyme sites in a number of genes in isolated nuclei. Surprisingly, our data indicate that there is no difference in the overall accessibility of sites for several restriction enzymes on active versus inactive genes. However, consistent with our model, restriction enzyme recognition sequences that overlap with homeoprotein binding sites are less accessible on inactive genes than they are on active genes. We propose that transcriptional activation in all animals may involve a localized increase in accessibility at the AT-rich regions bound by homeoproteins and perhaps at a few other regions, rather than a generalized effect on all sites throughout a gene.
INTRODUCTION
Sequence-specific DNA binding proteins are one of the largest groups of regulatory proteins in animals (1). Yet while the DNA binding specificities of these proteins have been studied extensively in vitro (2–4; http://transfac.gbf.de/transfac ), little information is available about the range of DNA sequences that most of these proteins interact with in cells. Even less is known about the mechanisms that determine the in vivo pattern of DNA binding. In vitro experiments have shown that cooperative interactions between sequence-specific DNA binding proteins can modify DNA binding specificities and that chromatin structure can inhibit DNA binding (5–9). But the relative contributions of these influences on DNA binding in animal cells are unknown. To address these issues, we are studying DNA binding by a collection of four Drosophila homeoproteins: Paired, Bicoid and two selector homeoproteins, Eve and Ftz.
Homeoproteins are a highly conserved family of transcription factors that control development in all animal phyla (10–12). Like other extended families of metazoan transcription factors, homeoproteins bind in vitro with overlapping specificities to variants of a short, degenerate DNA consensus sequence. For example, the selector homeoproteins all bind with similar specificities to variants of the consensus sequence NNATTA (13–15); Bicoid binds with high affinity to a smaller range of sequences, GGATTA, GCATTA, CGATTA and AGATTA (16–18); Paired binds not only to many ATTA sequences, but also to an entirely different 10–14 bp sequence via a second DNA binding domain contained within the Paired protein (19,20).
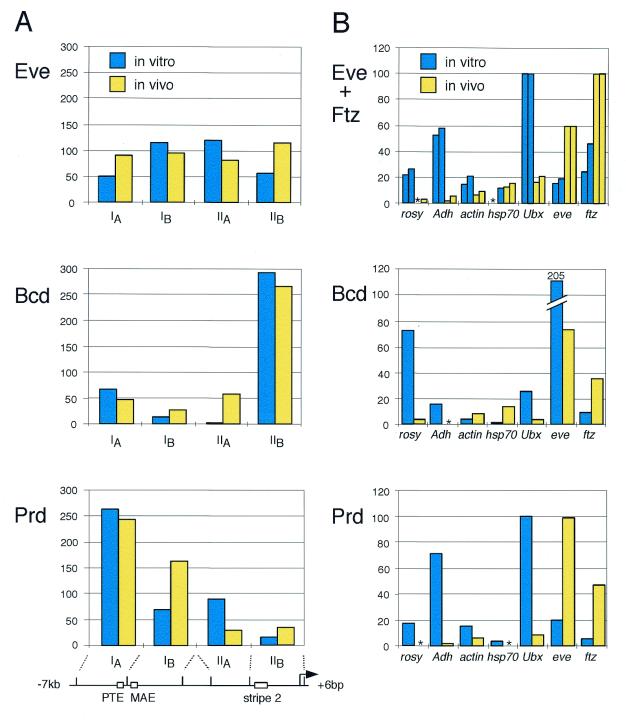
Using an in vivo UV crosslinking assay, we showed previously that Eve, Ftz, Paired and Bicoid have significantly overlapping DNA binding specificities in Drosophila embryos and that they bind to a broad array of genes (21,22). A detailed comparison of in vivo UV crosslinking data and in vitro DNA binding preferences suggest that, across the highly transcribed genes hb, eve and ftz, there is a strong correlation between the in vivo and in vitro DNA binding specificities of these proteins (Fig. 1A; 22,23). However, inactive or poorly transcribed genes, such as rosy, Adh and Ubx, are UV crosslinked significantly more weakly in vivo by all of these proteins than would be expected from the intrinsic affinities of homeoproteins for these DNA fragments in vitro (Fig. 1B).
Figure 1.
Comparison of the in vitro and in vivo DNA binding preferences of several Drosophila homeoproteins. (A) In vitro DNA binding and in vivo UV crosslinking across the eve promoter are broadly similar for Paired (Prd), Bicoid (Bcd) and Eve (21,22). The relative in vivo UV crosslinking (yellow) and in vitro binding (blue) per kb of DNA of each protein to four eve gene fragments (IA, IB, IIA and IIB) are shown. At the bottom is a diagram of the eve gene, indicating the relevant restriction fragments, the Paired target element (PTE), the minimal autoregulatory element activated by Eve (MAE), the Bicoid responsive stripe 2 element (stripe 2) and the mRNA start site (arrow). (B) In vitro DNA binding preferences differ from relative in vivo UV crosslinking when compared across differently transcribed genes. The relative in vivo UV crosslinking (yellow) and in vitro DNA binding (blue) per kb of DNA for four homeoproteins to a series of DNA fragments from several different gene loci are shown (21,22). In the panel showing data for Ftz and Eve, bars on the left of each pair show binding or crosslinking of Ftz and bars on the right show the equivalent data for Eve. The DNA restriction fragments shown range in size from 1.5 to 8 kb. An asterisk indicates that binding or crosslinking could not be detected. This figure was adapted from Carr and Biggin (22).
To explain these and other observations, we suggested that the pattern of homeoprotein DNA binding in vivo is determined by two principal factors (15,22). First, homeoproteins may be expressed at sufficiently high concentrations in vivo such that they bind efficiently to sites on actively transcribed genes; the chromatin structure of active genes is not envisioned to inhibit DNA binding and thus the intrinsic DNA binding specificities of homeoproteins should determine their distribution across the length of these genes. As Figure 1A demonstrates, this is apparently the case on the genes we have examined. Second, some structural feature may inhibit homeoprotein DNA binding at most sites on inactive or poorly transcribed genes. We suggest that this is why the relative binding on less active genes is lower in vivo than it is in vitro (Fig. 1B; rosy, Adh and Ubx).
In this paper, we have directly tested a key prediction of the above model. We have measured the relative accessibility of restriction sites in two highly transcribed and two weakly transcribed genes. Our results demonstrate that homeoprotein binding sites are indeed more accessible in highly transcribed genes than they are in weakly transcribed genes.
MATERIALS AND METHODS
Embryo UV irradiation assay
Drosophila embryos (stages 5–8) were collected and irradiated under 254 nm UV light for either 0 or 60 min using techniques described previously (21,24–26). DNA was extracted and purified also as previously described with the following changes. Isolated embryonic nuclei were treated with 100 µg/ml proteinase K for 5 min at 4°C and then lysed by the addition of sarkosyl to a final concentration of 4%. Restriction enzyme digests were carried out in a 100 µl volume of 1× restriction enzyme buffer (content specified by enzyme manufacturer) and contained 1 µg DNA from irradiated or unirradiated embryos and either 0, 60 or 120 U restriction enzyme. Reactions were allowed to proceed for 2 h at 37°C and were stopped by addition of proteinase K to 100 µg/ml. This was followed by incubation at 65°C for 30 min. The DNA was subsequently precipitated, washed and resuspended in standard loading buffer for separation on a 0.8% agarose gel. DNA was transferred to an uncharged nylon membrane and analyzed by Southern blot using standard procedures. Image data were quantitated with a Fuji Phosphorimager.
Restriction enzyme chromatin accessibility assay
Drosophila embryos (stages 5–8) were collected and dechorionated using standard techniques. Embryos were then suspended in buffer A (15 mM Tris pH 7.4, 60 mM KCl, 15 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.5 mM spermidine, 0.15 mM spermine, 0.5 mM DTT and 1 mM PMSF) at ∼1 g/3 ml and then immediately dissociated in a motorized dounce homogenizer. The homogenate was poured over Miracloth and diluted to 5 ml buffer A/g embryos. Embryos were then further homogenized with a B dounce homogenizer. This was followed by the addition of NP-40 to a final concentration of 0.5%. The released nuclei were then centrifuged at 4000 r.p.m. at 4°C for 15 min in a Sorval SS34 rotor. Pelleted nuclei were resuspended in an appropriate 1× restriction enzyme buffer (content specified by enzyme manufacturer) at 1 ml/g embryos and then centrifuged for 15 s in a microfuge. The nuclei were then resuspended in 1× restriction enzyme buffer (1 ml/g embryos) in preparation for restriction digestion. Nuclear chromatin digestion reactions were carried out for 30 min at 37°C in a 200 µl volume of 1× restriction enzyme buffer and contained 20 µl of resuspended nuclei and either 0, 100 or 200 U enzyme. Reactions were stopped by addition of proteinase K to 100 µg/ml and sarkosyl to 0.05%. After a 1 h incubation at 65°C, the DNA was phenol/chloroform extracted and ethanol precipitated. The DNA was then resuspended in 100 µl of 1× _Eco_RI restriction enzyme buffer with 0.05% Triton X-100 and digested for 2 h at 37°C with 100 U _Eco_RI. DNA was then precipitated and resuspended in standard loading buffer for separation on an agarose gel. Southern analysis was performed using standard techniques.
The following gene regions were examined: a 3.5 kb _Eco_RI fragment containing the ftz transcription unit; a 4.7 kb _Eco_RI fragment containing the Adh coding sequence; a 4.1 kb region extending 1.6 kb upstream and 2.5 kb downstream from the _Eco_RI site within the rosy gene; a 4.0 kb sequence bounded by _Eco_RI and _Bam_HI sites extending 3.8 kb upstream from the eve transcription start site. The following probes were used in the above assays: a 467 bp _Eco_RI–_Rsa_I ftz fragment located at the 5′-end of the above ftz encoding fragment; a 339 bp _Rsa_I–_Rsa_I ftz fragment located near the 3′-end of the above ftz encoding fragment; a 498 bp _Eco_RI–_Rsa_I Adh fragment located at the 5′-end of the above Adh gene fragment; a 646 bp _Eco_RI–_Rsa_I Adh fragment located at the 3′-end of the above Adh gene fragment; a 291 bp _Eco_RI–_Hae_III eve fragment that hybridizes to a sequence from 5.5 to 5.2 kb upstream of the eve transcription start site; a 322 bp _Eco_RI–_Rsa_I eve fragment that hybridizes to a sequence 7.8 to 7.5 kb upstream of the eve transcription start site; a 150 bp _Eco_RI–_Hae_III rosy downstream fragment; a 219 bp _Eco_RI–_Rsa_I rosy upstream fragment.
Primary restriction site digestion efficiencies were calculated with the formulae ADE = AINT, BDE = BINT/(1 – ADE), CDE = CINT/(1 – ADE)(1 – BDE), DDE = DINT/(1 – ADE)(1 – BDE)(1 – CDE), etc., where A is the primary restriction site at the end of the smallest restriction fragment and B is the primary restriction site at the end of the next largest fragment. DE is the digestion efficiency and INT is the intensity of each fragment band as a percentage of the total intensity in the gel lane. The digestion efficiencies of primary restriction sites that were too close together to resolve on the gel were averaged. For example, in the case of two unresolved primary restriction sites (BX and BY), the following was done. Assume BX DE = BY DE. Guesses of BX DE (and hence also of BY DE) are adjusted until BINT = BX INT + BY INT, where BX INT = BX DE(1 – ADE) and BY INT = BY DE(1 – BX DE) (1 – ADE). ADE is the digestion efficiency of the adjacent primary restriction site closer to the probed secondary restriction site and BINT is the intensity of the unresolved band. All image data were quantitated using a Fuji Phosphorimager.
It is formally possible that if different length DNA fragments transfer with different efficiencies in our Southern blots and if there is a systematic difference in the length of fragment produced by each enzyme on some genes, our results could be biased. However, as Table 1 indicates, the variations in the mean lengths of fragments transferred to Southern filters cannot explain the lower amounts of Adh and rosy DNA fragments produced by _Mse_I digestion (Fig. 4B). Further, the digestion efficiencies of a number of sites have been measured independently using partial digestion products that differ in length by 3-fold or more. In such cases, there is no significant difference in the amount of fragment produced (data not shown). Thus, the relative amount of each partial digestion product accurately reflects the relative digestion of the given restriction site in nuclei.
Table 1.
| Gene | Enzyme | Mean length of fragments measured on Southern blot |
|---|---|---|
| adh | _Mse_I | 1035 |
| adh | _Rsa_I | 1631 |
| adh | _Hae_III | 1399 |
| ftz | _Mse_I | 1880 |
| ftz | _Rsa_I | 1693 |
| ftz | _Hae_III | 1701 |
| eve | _Mse_I | 951 |
| eve | _Rsa_I | 960 |
| eve | _Hae_III | 733 |
| rosy | _Mse_I | 1103 |
| rosy | _Rsa_I | 2435 |
| rosy | _Hae_III | 1223 |
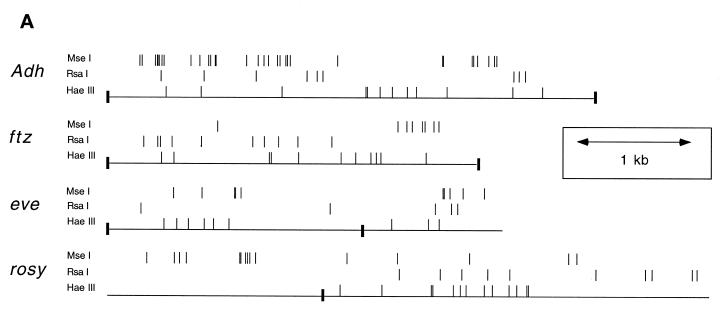
Figure 4.
_Mse_I restriction sites in eve and ftz are more heavily digested than those in Adh and rosy. (A) Restriction map indicating the positions of _Mse_I, _Rsa_I and _Hae_III sites in the Adh, ftz, eve and rosy genes. Only those sites whose accessibility to digestion was measured are shown. The scale in kb is indicated by the distance between the double-headed arrow. The positions of _Eco_RI sites are marked by filled rectangles. (B) Nuclei were isolated and then digested with either _Mse_I, _Rsa_I or _Hae_III. DNA was purified, digested to completion with a secondary enzyme and then separated on an agarose gel for Southern analysis. Blots were probed with fragments directed against either ftz, eve, Adh or rosy. Shown are the mean percent digestion for _Mse_I (yellow), _Rsa_I (purple) and _Hae_III (blue) sites across each of the above four genes. The error bars indicate 95% confidence limits for the mean.
Restriction enzyme digestion efficiencies of _Mse_I, _Rsa_I and _Hae_III sites were measured using a variety of other conditions, in addition to those described above. A number of the published nuclear preparation and restriction enzyme digestion methods were sampled, including those of Wu (27,28) and Wallrath (29). Higher or lower salt concentrations ranging from 0 to 60 mM KCl and 15 to 150 mM NaCl during preparation of nuclei or during primary restriction digestions were also tested, as was the presence of sucrose. All procedures gave similar data to those described in Results.
Statistical analyses
To compare the mean digestion efficiencies of _Mse_I sites on rosy and Adh with those on eve and ftz, the data for eve and ftz were first pooled. _F_-tests using an α of 0.05 indicate that the variances of the rosy and the pooled eve/ftz data can be considered equal, but that the variances of the Adh and the eve/ftz data are not equal. The appropriate _t_-tests were performed using Microsoft Excel. 95% confidence limits for the difference between each pair of means were calculated using the Excel derived _t_-statistic (7.12 for the Adh versus eve/ftz comparison and 3.34 for the rosy versus eve/ftz; 30). One tailed tests were appropriate as we had previously hypothesized that the digestion efficiencies of sites on rosy and Adh would be less than those on eve and ftz. The probability that the digestion efficiencies of _Mse_I sites on Adh are the same or greater than those on eve and ftz is 7.8 × 10–8. The probability that the digestion efficiencies of _Mse_I sites on rosy are the same or greater than those on eve and ftz is 1.1 × 10–3.
RESULTS
The eve and ftz genes are no more vulnerable to damage by UV irradiation than Adh and rosy
As described above, our idea that homeoprotein DNA binding is preferentially inhibited at poorly transcribed genes is based partly on the disparity between relative in vivo UV crosslinking and in vitro DNA affinity measurements on these genes (Fig. 1B). Our model assumes that in vivo UV crosslinking and in vitro DNA binding both accurately measure, in their respective environments, the relative occupancy of proteins on different gene fragments.
UV crosslinking and DNA affinity assays do give the same relative results when tested under the same conditions in vitro (22,23). It is crucial, however, to ensure that there is no factor(s) that interferes with the quantitative nature of the UV method in vivo. The only factor we can imagine that might have such an effect is if some genes receive different doses of UV irradiation than others. For example, less well transcribed genes may be physically buried within the nucleus and thus shielded from UV irradiation. In such a case, these genes would be UV crosslinked to homeoproteins at lower levels than highly transcribed genes because of their physical location and not, as our model assumes, because they are actually bound at lower levels. The eve and ftz genes are strongly transcribed at the stage of development examined in the in vivo UV crosslinking experiments, whereas Adh is not transcribed and rosy is only weakly transcribed in a subset of cells at the same stage. Therefore, we have sought to determine if the Adh and rosy genes receive the same amount of UV light as the eve and ftz genes during crosslinking experiments.
We have previously noted that, in DNA purified from UV irradiated embryos, some restriction enzyme sites in a small proportion of DNA fragments from each gene cannot be digested, even if the DNA has been treated with protease to remove attached proteins prior to restriction enzyme digestion (J.Walter, A.Carr, J.D.Laney and M.D.Biggin, unpublished data). Presumably, this inhibition of digestion is due to UV damage at restriction sites in some copies of the gene. The proportion of restriction sites that cannot be digested should thus be a measure of the amount of UV light incident at each gene. Restriction sites that contain two adjacent thymine residues, for example _Eco_RI sites, should be most susceptible to UV damage because thymine is more readily activated by UV light than are other bases and because adjacent thymines readily form thymine dimers upon activation (31). Therefore, to maximize our ability to detect damaged DNA sites, we have tested the susceptibility of _Eco_RI sites to UV light. To further increase our ability to measure UV damage, embryos have been UV irradiated for twice the time usual in standard UV crosslinking experiments.
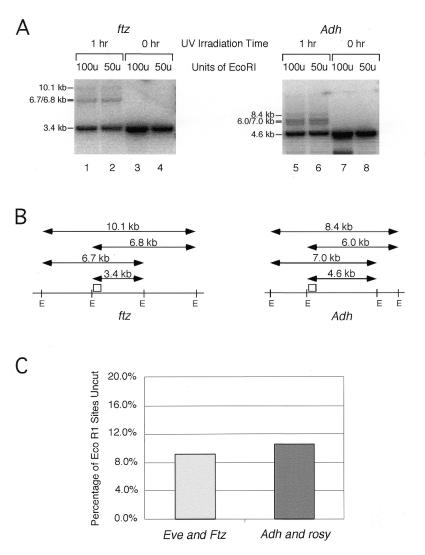
Live Drosophila embryos were irradiated with 254 nm light for either 0 or 60 min. DNA was extracted, proteinase K treated, purified, digested with _Eco_RI and then subjected to Southern analysis. As shown in Figure 2A, partial digestion products are observed in experiments using DNA from irradiated embryos, but not in experiments using DNA from unirradiated embryos. For example, a Southern blot probed with a short DNA fragment wholly contained within a 3.4 kb _Eco_RI ftz fragment detects only this 3.4 kb fragment in DNA from unirradiated embryos. But in DNA from irradiated embryos, larger DNA fragments of the size expected for partial digestion products are also visible (Fig. 2A and B). Digestion in the presence of higher concentrations of enzyme does not decrease the amount of partial digestion products observed (Fig. 2A). Also, if unirradiated plasmid DNA is included in a digest of irradiated DNA, an _Eco_RI site in the plasmid is completely digested (data not shown). Thus, the inhibition of DNA digestion must be due to physical obstruction on each DNA molecule that is not cut.
Figure 2.
The eve and ftz genes are no more accessible to UV light than the Adh and rosy genes in embryos. (A) Intact Drosophila embryos were collected and either UV irradiated for 1 h (lanes 1, 2, 5 and 6) or not irradiated (lanes 3, 4, 7 and 8). DNA was extracted, proteinase K treated and purified as described in Materials and Methods and then digested with either 100 (lanes 1, 3, 5 and 7) or 50 (lanes 2, 4, 6 and 8) U _Eco_RI. The DNA was then separated on an agarose gel and transferred to an uncharged nylon membrane for Southern analysis. Probes directed against an _Eco_RI restriction fragment from either ftz (left) or Adh (right) were used to detect the complete and partial digestion products shown. (B) The positions of the restriction fragments in (A) are indicated. Fully digested chromatin yields a 3.4 kb _Eco_RI fragment from the ftz loci and a 4.6 kb fragment from Adh. Incomplete digestion by _Eco_RI generates the larger restriction fragments shown. The DNAs used to probe the blots in (A) are indicated by small open boxes. (C) The average digestion efficiency of _Eco_RI sites in eve and ftz are compared to those in Adh and rosy. Three sites were examined in the rosy and Adh genes and four sites were examined in eve and ftz.
The mean proportion of _Eco_RI sites in Adh and rosy that cannot be digested in irradiated DNA is very similar to that of sites in eve and ftz (Fig. 2C). This result strongly suggests, therefore, that each of these four genes receives the same level of incident UV light during crosslinking experiments and that our in vivo UV crosslinking results must provide an accurate measure of the relative levels of binding of endogenous homeoproteins on these genes.
Restriction site accessibility in Drosophila embryonic nuclei
The above experiment confirms that there is a discrepancy between the in vitro affinities of homeoproteins for DNA and the in vivo levels of occupancy when genes transcribed to different degrees are examined (Fig. 1B). From this and other data, we earlier inferred that homeoprotein DNA binding sites should be less accessible on poorly transcribed genes than they are on strongly transcribed genes (15,22). Below, we use a restriction enzyme accessibility assay to directly test this idea.
It has previously been shown that, in isolated nuclei, most DNA sites in SIR silenced yeast genes are 10 times less accessible to restriction digestion than are sites in the same genes when they are active (32). In other yeast genes and in some metazoan genes, short regions of DNA just upstream of the mRNA start site become much more accessible to restriction enzyme digestion when these genes are activated (33–35). This assay may thus provide a useful way to test our model.
We have compared the digestion efficiency of restriction sites within eve and ftz to those within rosy and Adh in isolated nuclei. We measured the accessibility of three enzymes: _Rsa_I, _Hae_III and _Mse_I. _Hae_III and _Rsa_I were chosen because their recognition sequences, GGCC and GTAC, respectively, are very different from those of homeoproteins. Across the restriction fragments we have examined, only 1 of 40 _Rsa_I sites (2.5%) and 9 of 75 _Hae_III sites (12%) are within 10 bp of a copy of the homeoprotein core recognition sequence, ATTA. Thus, these enzymes allow us to measure the general accessibility of each gene locus at sites not bound by homeoproteins. _Mse_I was chosen because it cuts the sequence TTAA. This sequence is a 1 bp mismatch from the homeoprotein core recognition sequence and, as a consequence, many homeoprotein DNA binding sites contain an _Mse_I site. Within the genes we have analyzed, 52 of 80 _Mse_I sites (65%) either overlap or are within 10 bp of an ATTA element. In addition, because homeoproteins bind strongly to some 1 bp mismatches to ATTA, many of the other 35% of _Mse_I sites are within or close to sequences predicted to be bound by homeoproteins. This enzyme should thus allow us to monitor accessibility at homeoprotein binding sites.
Accessibility was measured by indirect end-labeling (27,29). Briefly, nuclei were purified from Drosophila embryos of developmental stage 4–5, the same stage as used in the in vivo UV crosslinking experiments. These nuclei were then immediately added to a restriction enzyme solution containing either _Rsa_I, _Hae_III or _Mse_I (Fig. 3A). This primary digestion continued until all accessible restriction sites had been cut: digestion of nuclei for longer or with more enzyme gave no further cutting (data not shown). After digestion, the DNA was purified from nuclei, removing all histones and other proteins from the DNA. Then, the DNA was cut with a secondary restriction enzyme, which recognizes a 6 bp sequence. In this digestion, all sites recognized by the secondary enzyme were cut to completion, delimiting specific gene regions for analysis. The double-digested DNA was then separated on an agarose gel and transferred to a nylon membrane for Southern analysis. Blots were probed with a short DNA fragment from one end of a length of DNA bounded by secondary digestion sites (Fig. 3B). Each fragment detected by a probe thus shares a common secondary enzyme site at one end and has a unique primary restriction enzyme site at the other end. The relative intensities of each fragment were used to quantitate the digestion efficiency of each primary restriction site (Materials and Methods). Note that there is no systematic difference between the size of fragments produced by partial digestion of _Mse_I, _Rsa_I and _Hae_III that could bias our analysis (Materials and Methods).
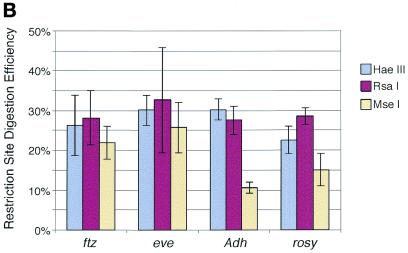
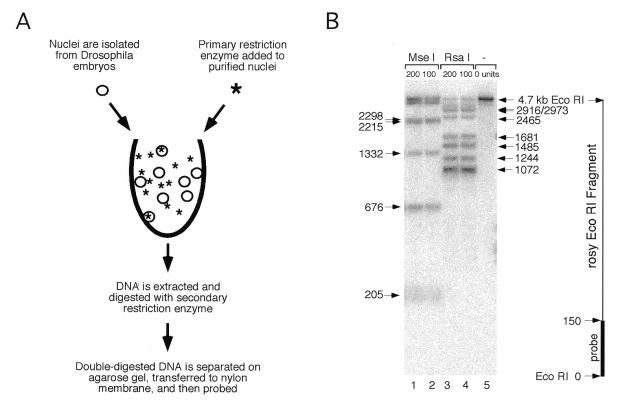
Figure 3.
The restriction site accessibility assay. (A) Nuclei were isolated from live Drosophila embryos and incubated with a primary restriction enzyme for 30 min at 37°C. DNA was then purified from nuclei and digested to completion with a secondary restriction enzyme. The DNA fragments were separated on an agarose gel and transferred to a nylon membrane in preparation for Southern analysis. (B) An example of a Southern blot showing partial digestion of sites for two primary enzymes in the rosy gene. DNA was extracted from nuclei that had been treated with either _Mse_I (lanes 1 and 2), _Rsa_I (lanes 3 and 4) or no enzyme (lane 5) and then digested with a secondary enzyme, _Eco_RI. A 150 bp probe directed against sequences adjacent to an _Eco_RI restriction site near the 3′-end of the rosy transcription unit was used to detect _Rsa_I and _Mse_I restriction fragments generated during the primary restriction enzyme digestion of nuclei. The nucleotide positions of _Mse_I sites are indicated to the left and those of _Rsa_I sites on the right. A map of the 4.7 kb _Eco_RI rosy fragment is shown on the far right.
The digestion efficiencies of sites for _Mse_I, _Rsa_I and _Hae_III were calculated and averaged across rosy, Adh, eve and ftz (Fig. 4). Because some primary restriction sites are either too close or too far from secondary restriction sites, it was not possible to measure accessibility at all sites. Data was obtained for 83% of _Mse_I sites, 85% of _Rsa_I sites and 60% of _Hae_III sites within the designated regions (Fig. 4A). The histogram indicates 95% confidence limits for the mean digestion of each enzyme on each DNA (Fig. 4B). As the figure shows, there is no significant difference in average digestion efficiency of _Rsa_I and _Hae_III at the four genes examined. However, the average digestion efficiency of _Mse_I sites is lower on rosy and Adh than it is on eve and ftz. One tailed _t_-tests indicate that with 95% confidence the _Mse_I sites on Adh are digested between 1.8- and 2.4-fold less well than they are on eve and ftz and the _Mse_I sites on rosy are digested 1.3- to 1.9-fold less well than they are on eve and ftz (Materials and Methods). We obtain essentially the same result if we compare just those _Mse_I sites that overlap or lie within 10 bp of ATTA elements. Thus, consistent with our model, the DNA sequences in and around homeoprotein binding sites on Adh and rosy are less accessible than comparable sites on the eve and ftz genes. The fact that homeoprotein binding sites on Adh are less accessible than those on rosy is also consistent with our model: at the stage of development studied in the in vivo UV crosslinking experiments, Adh is not transcribed in any cells, whereas rosy is weakly transcribed in a minority of cells. We discuss the implications of these data below.
DISCUSSION
In vivo homeoproteins bind more weakly to poorly transcribed genes than predicted by in vitro DNA binding data (Fig. 1; 22,23). We have shown that UV crosslinking gives an accurate and quantitative measure of the relative levels of protein binding to different affinity DNA fragments (22–24) and, in this paper, we establish that the same amount of UV light reaches all genes in Drosophila embryos. Therefore, the in vitro and in vivo DNA binding specificities of homeoproteins differ.
In principle, there are two ways that DNA binding specificities could be altered in vivo. One, DNA binding could be specifically increased at actively transcribed genes by heteromeric cooperative interactions between homeoproteins and other sequence-specific DNA binding proteins. Two, DNA binding by homeoproteins could be inhibited at weakly transcribed genes. The following data suggest that the second of these two mechanisms is the principal means by which homeoprotein DNA binding is modified in vivo.
A variety of experiments suggest that homeoproteins bind to high affinity DNA sites on actively transcribed genes without the aid of cooperative interactions with other sequence-specific DNA binding proteins (15,22,36). For example, the in vitro and in vivo DNA binding preferences of homeoproteins are very similar on actively transcribed genes (Fig. 1A). Yet if, as the first model suggests, cooperative interactions with other sequence-specific DNA binding proteins greatly affect homeoprotein DNA binding in vivo, the in vitro and in vivo specificities would differ on actively transcribed genes. As a second example, transgenic promoter constructs containing only high affinity Bicoid recognition sequences are activated by Bicoid in embryos (37,38). Thus, in some contexts, endogenous Bicoid does not need to form heteromeric complexes with other transcription factors to bind its recognition sites in vivo. By default, if homeoprotein DNA binding is not greatly increased on actively transcribed genes, the major factor modifying DNA binding in vivo must be the inhibition of binding to inactive genes.
To directly test this idea, we have used a restriction enzyme accessibility assay. This assay demonstrates that sites bound by homeoproteins are 1.6- to 2.2-fold less accessible on poorly transcribed genes than they are on actively transcribed genes (Fig. 4). Thus, two separate experimental approaches both indicate that homeoprotein DNA binding is differentially inhibited on less well transcribed genes.
Perhaps surprisingly, we do not see differences in accessibility on active versus inactive genes at sites cut by the restriction enzymes _Rsa_I and _Hae_III, which do not recognize homeoprotein DNA binding sites (Fig. 4). It is unlikely that this is because different restriction enzymes respond differently to altered chromatin structures; previous studies have not found such effects and, instead, have shown close agreement between DNase I, micrococcal nuclease and restriction enzyme mapping data (33). Therefore, our results suggest that upon activation of Drosophila genes, accessibility is increased locally at regions around homeoprotein DNA sites. A general increase in accessibility throughout the length of genes, such as that associated with derepression of SIR silencing in yeast, is not observed (32,39); though, from our data, we cannot rule out that a few other regions, in addition to homeoprotein DNA sites, may display increased accessibility upon gene activation.
Typically, homeoprotein binding sites are present as clusters that form short AT-rich patches in the non-protein coding portions of Drosophila genes (13,40; T.Johnson, D.Dalma-Weiszhausz, M.D.Biggin and M.Gerstein, unpublished data). These patches might lie within the center of nucleosomes on inactive genes but may be placed at the edge or between nucleosomes on active genes. Such differential positioning of nucleosomes can cause up to 10-fold differences in accessibility (41), which could explain the differential accessibility we observe. Interestingly, the human SWI–SNF chromatin remodeling complex contains a HMG I/Y-like DNA binding domain that shows a high preference for AT-rich DNA (42); and it is thought that SWI–SNF complexes are specifically recruited to transcriptionally active genes (6–8). These complexes, therefore, may be found most frequently at AT-rich patches within active genes and preferentially increase accessibility at these sites. Consistent with this idea, hypomorphic mutations in components of the Drosophila SWI–SNF-like remodeling complexes often have specific effects on homeoprotein gene activity (43–45). While these remodeling complexes probably have broad effects on DNA binding by many classes of transcription factor, they may play a more important role for the homeoproteins.
Although the restriction enzyme accessibility data agrees qualitatively with the predictions resulting from the comparison of in vitro and in vivo DNA binding (Fig. 1), the two experimental approaches give different estimates of the degree to which homeoprotein binding is inhibited on inactive genes. Given the disparity between the relative in vitro and in vivo DNA binding preferences of homeoproteins, DNA sites in the eve and ftz genes should be about 10 times more accessible than sites in the Adh and rosy genes in blastoderm embryos (see Fig. 1). However, the restriction enzyme assay shows only a 1.6- to 2.2-fold mean reduction in accessibility at rosy and Adh compared to eve and ftz (Fig. 4). We suspect that the reason for this difference is that the enzyme accessibility assay may be underestimating the true difference in accessibility in vivo.
There are a number of reasons why the present assay may not be fully quantitative. For example, during the isolation of nuclei, some aspects of in vivo structure may be lost. Also, sequence-specific DNA binding proteins may block accessibility of restriction enzymes to their recognition sites, though perhaps to a lesser extent than the presence of nucleosomes. Given that most homeoprotein binding sites are likely to be occupied by homeoproteins on the eve and ftz genes when they are actively transcribed, the restriction enzyme assay may underestimate the accessibility of these sites for homeoproteins. Indeed, the average extent to which homeoprotein recognition sites are cut in eve and ftz is only ∼30% (Fig. 4) and no homeoprotein sites are cut by >40%. Thus, the true degree of accessibility of transcription factor sites is inherently difficult to measure with this method. We suggest that it will be important to develop new approaches to accurately quantitate DNA accessibility in cells in the future.
Acknowledgments
ACKNOWLEDGEMENTS
We are indebted to Lori Wallrath for advice on the restriction enzyme accessibility assay and for crucial insights. We thank Trevor Williams, Tony Koleske and Janet Carr for helpful comments on this manuscript. This work was funded in part by a grant from the National Institutes of Health to M.D.B.
REFERENCES
- 1.Ruvkun G. and Hobert,O. (1998) Science, 282, 2033–2041. [DOI] [PubMed] [Google Scholar]
- 2.Faisst S. and Meyer,S. (1992) Nucleic Acids Res., 20, 3–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pabo C.O. and Sauer,R.T. (1992) Annu. Rev. Biochem., 61, 1053–1095. [DOI] [PubMed] [Google Scholar]
- 4.Heinemeyer T., Wingender,E., Reuter,I., Hermjakob,H., Kel,A.E., Kel,O.V., Ignatieva,E.V., Ananko,E.A., Podkolodnaya,O.A., Kolpakov,F.A., Podkolodny,N.L. and Kolchanov,N.A. (1998) Nucleic Acids Res., 26, 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mann R.S. and Chan,S.K. (1996) Trends Genet., 12, 258–262. [DOI] [PubMed] [Google Scholar]
- 6.Workman J.L. and Kingston,R.E. (1998) Annu. Rev. Biochem., 67, 545–579. [DOI] [PubMed] [Google Scholar]
- 7.Kornberg R.D. and Lorch,Y. (1999) Curr. Opin. Genet. Dev., 9, 148–151. [DOI] [PubMed] [Google Scholar]
- 8.Kadonaga J.T. (1998) Cell, 92, 307–313. [DOI] [PubMed] [Google Scholar]
- 9.Scott M.P. (1999) Nature, 397, 649–651. [DOI] [PubMed] [Google Scholar]
- 10.Burglin T.R. (1994) In Duboule,D. (ed.), Guidebook to the Homeobox Genes. Oxford University Press, Oxford, UK, pp. 27–269.
- 11.Finnerty J.R. and Martindale,M.Q. (1998) Curr. Opin. Genet. Dev., 8, 681–687. [DOI] [PubMed] [Google Scholar]
- 12.de Rosa R., Grenier,J.K., Andreeva,T., Cook,C.E., Adoutte,A., Akam,M., Carroll,S.B. and Balavoine,G. (1999) Nature, 399, 772–776. [DOI] [PubMed] [Google Scholar]
- 13.Desplan C., Theis,J. and O’Farrell,P.H. (1988) Cell, 54, 1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gehring W.J., Affolter,M. and Burglin,T. (1994) Annu. Rev. Biochem., 63, 487–526. [DOI] [PubMed] [Google Scholar]
- 15.Biggin M.D. and McGinnis,W. (1997) Development, 124, 4425–4433. [DOI] [PubMed] [Google Scholar]
- 16.Driever W. and Nusslein-Volhard,C. (1989) Nature, 337, 138–143. [DOI] [PubMed] [Google Scholar]
- 17.Percival-Smith A., Muller,M., Affolter,M. and Gehring,W.J. (1990) EMBO J., 9, 3967–3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Small S., Blair,A. and Levine,M. (1992) EMBO J., 11, 4047–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujioka M., Miskiewicz,P., Raj,L., Gulledge,A.A., Weir,M. and Goto,T. (1996) Development, 122, 2697–2707. [DOI] [PubMed] [Google Scholar]
- 20.Jun S. and Desplan,C. (1996) Development, 122, 2639–2650. [DOI] [PubMed] [Google Scholar]
- 21.Walter J., Dever,C.A. and Biggin,M.D. (1994) Genes Dev., 8, 1678–1692. [DOI] [PubMed] [Google Scholar]
- 22.Carr A. and Biggin,M.D. (1999) EMBO J., 18, 1598–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walter J. and Biggin,M.D. (1996) Proc. Natl Acad. Sci. USA, 93, 2680–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walter J. and Biggin,M.D. (1997) Methods, 11, 215–224. [DOI] [PubMed] [Google Scholar]
- 25.Carr A. and Biggin,M. (1998) In Becker,P.B. (ed.), Methods in Molecular Biology. Humana Press, Totowa, NJ, p. 119.
- 26.Biggin M.D. (1999) Methods Enzymol., 304, 496–515. [DOI] [PubMed] [Google Scholar]
- 27.Wu C., Wong,Y.C. and Elgin,S.C. (1979) Cell, 16, 807–814. [DOI] [PubMed] [Google Scholar]
- 28.Wu C. (1984) Nature, 309, 229–234. [DOI] [PubMed] [Google Scholar]
- 29.Wallrath L.L., Swede,M.J. and Elgin,S.C.R. (1998) In Gould,H. (ed.), Chromatin A Practical Approach. Oxford University Press, Oxford, UK.
- 30.Anderson T.W. and Finn,J.D. (1996) The New Statistical Analysis of Data. Springer-Verlag, New York, NY.
- 31.Nikogosyan D.N. (1990) Int. J. Radiat. Biol., 57, 233–299. [DOI] [PubMed] [Google Scholar]
- 32.Loo S. and Rine,J. (1994) Science, 264, 1768–1771. [DOI] [PubMed] [Google Scholar]
- 33.Almer A., Rudolph,H., Hinnen,A. and Horz,W. (1986) EMBO J., 5, 2689–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wallrath L.L. and Elgin,S.C. (1995) Genes Dev., 9, 1263–1277. [DOI] [PubMed] [Google Scholar]
- 35.Beato M. and Eisfeld,K. (1997) Nucleic Acids Res., 25, 3559–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X., Murre,C. and McGinnis,W. (1999) EMBO J., 18, 198–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanes S.D., Riddihough,G., Ish-Horowicz,D. and Brent,R. (1994) Mol. Cell. Biol., 14, 3364–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simpson-Brose M., Treisman,J. and Desplan,C. (1994) Cell, 78, 855–865. [DOI] [PubMed] [Google Scholar]
- 39.Gottschling D.E. (1992) Proc. Natl Acad. Sci. USA, 89, 4062–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Appel B. and Sakonju,S. (1993) EMBO J., 12, 1099–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kladde M.P. and Simpson,R.T. (1996) Methods Enzymol., 274, 214–233. [DOI] [PubMed] [Google Scholar]
- 42.Bourachot B., Yaniv,M. and Muchardt,C. (1999) Mol. Cell. Biol., 19, 3931–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elfring L.K., Daniel,C., Papoulas,O., Deuring,R., Sarte,M., Moseley,S., Beek,S.J., Waldrip,W.R., Daubresse,G., DePace,A., Kennison,J.A. and Tamkun,J.W. (1998) Genetics, 148, 251–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crosby M.A., Miller,C., Alon,T., Watson,K.L., Verrijzer,C.P., Goldman-Levi,R. and Zak,N.B. (1999) Mol. Cell. Biol., 19, 1159–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daubresse G., Deuring,R., Moore,L., Papoulas,O., Zakrajsek,I., Waldrip,W.R., Scott,M.P., Kennison,J.A. and Tamkun,J.W. (1999) Development, 126, 1175–1187. [DOI] [PubMed] [Google Scholar]