Expression of the Human Endogenous Retrovirus HTDV/HERV-K Is Enhanced by Cellular Transcription Factor YY1 (original) (raw)
Abstract
The human endogenous retrovirus HTDV/HERV-K, which resides in moderate copy numbers in the human genome, is expressed in a cell-type-specific manner, predominantly in teratocarcinoma cells. We have analyzed the regulatory potential of the 5′ enhancer of the HERV-K long terminal repeat. Protein extracts of HERV-K-expressing teratocarcinoma cell lines (GH and Tera2) and nonexpressing HeLa and HepG2 cells form different protein complexes on the enhancer sequence as detected by electrophoretic mobility shift assays (EMSA). Using competition EMSAs, DNase I footprinting, and supershift experiments, we localized the binding site of these complexes to a 20-bp sequence within the enhancer and showed that the transcription factor YY1 is one component of the HERV-K enhancer complex. Replacement of the YY1 binding site with unrelated sequences reduced expression of the luciferase gene as a reporter in transient-transfection assays.
Endogenous retroviral sequences comprise a significant proportion of the genome in all vertebrates. They probably are remnants of ancient germ line infections of exogenous retroviruses which have been transmitted as stable genetic traits via the germ line. Thus, endogenous retroviruses (ERVs) in principle have the same genomic organization as exogenous ones. Approximately 1 to 12% of the human genome consists of ERV elements. The high copy number most probably arose by repeated cycles of infection and retrotransposition. ERVs may still significantly contribute to recombination events and genetic instability. Many different families of ERVs have been detected in the human genome. However, the vast majority of them have accumulated point mutations and deletions over time, leaving either full-length or truncated proviruses, which are unable to code for infectious virus particles (reviewed in references 21 and 41). Almost all endogenous retroviruses are expressed on the RNA level in a rather cell-type-specific manner; however, only a few of them encode functional proteins. In agreement with their fate as retroelements, ERVs are transcribed primarily in cells and tissues of the reproductive tract or in dedifferentiated tumor cells (14, 21, 41).
The human endogenous retrovirus HTDV/HERV-K family consists of 25 to 50 proviral copies and approximately 10,000 solitary long terminal repeats (LTRs), which presumably originated from homologous LTR-LTR recombination and excision of the retroviral genome in between (16, 26). HERV-K proviruses have retained the full complement of retroviral genes (gag, pol, and env) and additionally contain a central open reading frame encoding a small protein, termed cORF, with similarities to the HIV Rev protein (20). Homologs of HERV-K are found only in Old World monkeys and other primates, so that the primary germ line infection is traced back to a time after the separation of these species from New World monkeys (36). Despite this long period, the retroviral genes of HERV-K have retained coding competence, leading to formation of all viral protein products (reviewed in references 21 and 40). However, it is not known whether different proviruses complement intact genes in trans or whether a single provirus contains all the retroviral genes in one functional sequence. HERV-K is highly expressed in teratocarcinoma cell lines like GH and Tera2, where four mRNA species have been identified: a full-length mRNA (8.6 kb), a singly spliced env mRNA (3.3 kb), a doubly spliced cORF mRNA (1.8 kb), and a further singly spliced mRNA (1.5 kb) of unknown function. This expression pattern thus resembles that of exogenous retroviruses with complex regulation (18–20). Moreover, due to its coding competence, HERV-K is the only ERV known to encode retroviral particles, termed human teratocarcinoma-derived virus particles (HTDV); however, they seem not to be infectious (4, 17, 19).
Expression of retroviral sequences is regulated primarily by transcriptional regulatory elements within the LTR, especially the U3 region, at the 5′ end of the provirus. Cellular factors bind to these elements to initiate transcription (22). LTRs not only regulate the expression of their own downstream proviruses but also may influence the expression of neighboring cellular genes. The 3′ LTR of the mouse intracisternal A particles (IAPs) initiates transcription of the homeobox gene Hox-2.4 (12). The human HERV-R provirus generates a readthrough hybrid transcript from its 5′ LTR, with a downstream gene (H-plk) encoding a putative zinc finger protein (9). A hybrid transcript between HERV-H and the cellular gene PLA2L, which contains two phospholipase A2 homologous domains, is also known (7). Moreover, during primate evolution, insertion of a HERV-E element in front of an ancestral amylase gene converted its pancreas-specific promoter to a parotidic promoter as well. In this case, retroviral insertion altered the tissue-specific expression of a downstream cellular gene (39). So far, no cellular gene is known to be regulated by HERV-K; however, given the large number of HERV-K LTR copies in the human genome, it seems likely that many examples await discovery. The above examples clearly stress the importance of studying the potential of endogenous LTRs as regulatory elements of cellular genes.
The U3 region of many retroviruses contains two major domains which control expression: a promoter immediately preceding the transcription start site at the U3-R boundary and an enhancer domain further upstream within U3 (22). Detailed studies have been performed to reveal the regulation of expression of exogenous retroviruses like the avian retroviruses or human immunodeficiency virus (e.g., see references 10 and 30); however, much less is known for ERVs. Transcription of ERV-9, for example, is initiated at its promoter by the cellular transcription factor Sp1 and an unidentified protein binding to an initiator-like element (13, 37). Members of the Sp1 family also activate transcription from the HERV-H promoter (35). For HERV-K, we have identified a 5′ enhancer at the very 5′ end of U3 and a minimal promoter region at its 3′ end by analyzing the ability of LTR deletion constructs to drive the expression of the luciferase gene as a reporter in transient-transfection experiments with teratocarcinoma cells. Deletion of either the enhancer or the promoter markedly reduced the expression of the reporter, indicating that both regions are important determinants of transcriptional activation from the HERV-K LTR (11).
In this study, we have further analyzed the HTDV/HERV- K 5′ enhancer. Using electrophoretic mobility shift assays (EMSA), we detected different protein complexes in nuclear extracts of HERV-K-expressing cell lines (GH and Tera2), as opposed to nonexpressing ones (HeLa and HepG2). All complexes assemble at only a short sequence between bp 62 and 83 and contain the cellular transcription factor YY1 as a major DNA-binding protein. In luciferase reporter assays, we further show that these 5′-enhancer complexes activate HERV-K expression.
MATERIALS AND METHODS
Cell culture.
The human teratocarcinoma cell lines GH and Tera2 (American Type Culture Collection) (17), the human hepatocarcinoma cell line HepG2, and the human cervical carcinoma cell line HeLa (both purchased from the American Type Culture Collection) were grown at 37°C in Dulbecco’s modified Eagle’s medium (Biochrom) supplemented with 10% fetal calf serum, 2 mM glutamine, and antibiotics. The cells were split weekly after trypsinization in a 1:10 (GH, Tera2, and HepG2) or 1:20 (HeLa) ratio, with one change of medium after 3 days.
Protein preparation.
Protein extracts were prepared as described previously (6) and stored under a nitrogen atmosphere at −70°C. The protein concentration was measured by the Bio-Rad protein assay.
Cloning of the LTR subfragments.
The following oligonucleotides (Eurogentec, Interactiva) were used for cloning; the name, sequence, and approximate position within the HERV-K10 LTR are given (positions according to reference 26); mutated bases are in boldface type, restriction sites are underlined, and the respective restriction enzyme is mentioned: MK13, GAGAGATCGAATTCTTACTGTG, nucleotide (nt) 25, _Eco_RI; MK14, TAACAGAATCTCGAGGCAGAAG, nt 105, _Xho_I; MK15, GACTCCATCTAGATATGTGCTAAG, nt 75, _Xba_I; MK16, GAGCACGGAATTCGGGGTAAGGTC, nt 135, _Eco_RI; MK18, CATTCAACCTCGAGTTGACACAGC, nt 170, _Xho_I; MK21, P- AGCTTAGACATAGGAGACTCCATTA, nt 60, _Hin_dIII; MK22, P-AGCTTAATGGAGTCTCCTATGTCTA, nt 65, _Hin_dIII; MK23, P- AGCTTAGGAGACTCCATTTTGTTATGTGCTA, nt 70, _Hin_dIII; MK24, P- AGCTTAGCACATAACAAAATGGAGTCTCCTA, nt 75, _Hin_dIII; MK25, P- AGCTTGTGTAGAAAGAAGTAGACATAGGA, nt 50, _Hin_dIII; MK26, P-AGCTTCCTATGTCTACTTCTTTCTACACA, nt 55, _Hin_dIII; MK27, P- AGCTTTTTGTTATGTGCTAAGAAAAA, nt 80, _Hin_dIII; MK28, P-AGCTTTTTTCTTAGCACATAACAAAA, nt 85, _Hin_dIII; MK29, P- AGCTTAAGAAAAATTCTTCTGCCTTGAGA, nt 95, _Hin_dIII; MK30, P-AGCTTCTCAAGGCAGAAGAATTTTTCTTA, nt 100, _Hin_dIII; MK39, CCCGGGCTGCAGGAATTCGATATCATGTCTACTTCTTTCTAC, nt 60; MK40, GATATCGAATTCCTGCAGCCCGGGTAAGAAAAATTCTTCTGCC, nt 80; MK41, CGGTATCGATAAGCTTTGTGGGGAAAAGCAAGAG, nt 10, _Cla_I, _Hin_dIII; and MK42, GGGAGAAACCTTGGACAATACCTGG, nt 340.
The plasmids used in this work are based on pBHK10LTR, which contains the full-length HERV-K10 LTR (unpublished data). HERV-K10 LTR subfragments were amplified by PCR from pBHK10LTR with Taq DNA polymerase (Perkin-Elmer) and the primers listed above. Subfragments were restricted according to the restriction sites introduced by the primers, purified by agarose gel electrophoresis on low-melting-point (LMP) agarose, and ligated into appropriately restricted and dephosphorylated pBluescript (Stratagene). Recombinant plasmids were transformed into Escherichia coli DH5α (Gibco BRL) and isolated by using a plasmid kit (Qiagen) as specified by the manufacturer. All plasmids were verified by sequencing with the Sequenase V2.0 DNA sequencing kit (Amersham).
To produce pB30-103K10. The 102-bp PCR product, amplified with MK13 and MK14, was restricted with _Eco_RI and _Xho_I and ligated into pBluescript, which had been digested with the same enzymes. The same strategy was also used the next four constructs, so only the primers and enzymes used to produce the PCR fragments along with their sizes are mentioned: pB30-134K10, MK13/MK16, 132-bp product, and _Eco_RI; pB78-170K10, MK15/MK18, 119-bp product, and _Xba_I und _Xho_I; and subfragments [1–54] and [1–167], _Hin_dIII-_Acc_I and _Hin_dIII-_Hin_cII restriction fragments of the HERV-K10 LTR, respectively. After digestion of the LTR, the fragments were purified by agarose gel electrophoresis on LMP agarose.
To produce the following constructs, 5′-phosphorylated oligonucleotides (listed above) were annealed to yield double-stranded DNA fragments, which contained _Hin_dIII sites at both ends and the HERV-K10 LTR sequence as demarcated in the plasmid name (in base pairs). The annealed fragments then were cloned into _Hin_dIII-digested, dephosphorylated pBluescript: pB40-64K10, MK25/MK26 hybrid; pB54-73K10, MK21/MK22 hybrid; pB60-86K10, MK23/MK24 hybrid; pB72-93K10, MK27/MK28 hybrid; pB85-109K10, MK29/MK30 hybrid. All subfragments were isolated after _Hin_dIII digestion and electrophoresis with LMP agarose gels as described above.
For the YY1 binding-site mutant construct pB60PL85HK10, the LTR region from bp −16 through 84 (containing 16 bp of the pBluescript polylinker 5′ of the LTR, including a _Cla_I site and a _Hin_dIII site) and the region from bp 61 through 350 (including a _Sph_I site) were amplified by PCR from pBHK10LTR with primers MK41 plus MK39 and MK40 plus MK42, respectively. Both overlapping PCR products were used as templates in an assembly PCR with primers MK41 and MK42 to produce the 5′-terminal 350 bp of the LTR containing the mutant YY1 binding site. The _Cla_I- and _Sph_I-restricted product was used to substitute the _Cla_I-_Sph_I fragment of the LTR in plasmid pBHK10LTR to create a full-length LTR containing the mutated binding site. The mutant LTR was isolated by _Hin_dIII digestion and cloned into the _Hin_dIII-cut luciferase vector pSVOAL (5).
EMSA and supershift assay.
Plasmid pBHK10, containing the full-length HERV-K10 LTR cloned into the _Hin_dIII site of pBluescript, was restricted with _Hin_dIII and _Hin_cII to generate the LTR fragment bp 1 to 167. The other subfragments were excised from the subcloned constructs listed above by _Hin_dIII digestion. The calf intestinal phosphatase (Promega)-dephosphorylated DNA probes were separated by electrophoresis in LMP agarose gels (Biozym) and visualized by ethidium bromide staining. Probe bands were cut out, supplemented with 115 mM NaCl and 11.75 mM EDTA, extracted from melted gel slices with phenol (once), phenol-chloroform (once), and chloroform (twice), and precipitated with ethanol containing 10 mM MgCl2. A 50-ng portion of probe DNA was end labeled with T4 polynucleotide kinase (Promega) by using 10 μCi of [γ-32P]ATP (3,000 Ci/mmol; Amersham). Labeled probes were purified from unincorporated nucleotides by chromatography on a Sephadex G-50 column (Pharmacia). A specific activity of 2 × 107 to 8 × 107 cpm/μg was routinely obtained.
A protein binding reaction mixture consisting of 0.09 to 0.12 ng of probe DNA (6,000 to 15,000 cpm), 10 mM Tris-HCl (pH 7.5), 50 mM NaCl, 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol, 4% glycerol, 1 μg of poly(dI-dC) and 2 to 3 μg of nuclear protein extract in a total volume of 20 μl was incubated for 30 min at room temperature. In competition EMSAs, the reaction mixture was preincubated for 20 min with a 400- to 500-fold excess of competitor DNA before addition of labeled probe. In supershift assays, the binding-reaction mixture was supplemented with 1.2 μg of anti-YY1 immunoglobulin G (Santa Cruz Biotechnology) or 1 μl of cORF antiserum (20). The reaction was stopped by adding 2 μl of 10× loading buffer (250 mM Tris-HCl [pH 7.5], 0.2% bromphenolblue, 0.2% xylene cyanol, 40% glycerol), and the product was immediately loaded on a native 5% (4% for supershifts) polyacrylamide gel (5% acrylamide, 0.26% bisacrylamide [Gel 40, 19:1; Roth], 0.5× Tris-borate-EDTA [TBE], 2.5% glycerol). Electrophoresis was performed in a Protean II xi system (Bio-Rad) in 0.5× TBE buffer at 10 V/cm; the gel was prerun at 15 V/cm for 20 min. The gel was blotted on filter paper (3 MM; Whatman), dried at 80°C, and exposed overnight at −70°C with a Biomax MR film (Kodak).
DNase I footprinting.
The full-length HERV-K10 LTR (_Hin_dIII restriction fragment of pBHK10) was 5′-end labeled as described above and restricted with _Hin_cII to generate the LTR fragment from bp 1 to 167, which was radioactively labeled only at one end (at bp 1). The probe was purified from LMP agarose (see above). Less than 1 ng of probe DNA was incubated in EMSA buffer with 6 to 8 μg of nuclear protein extract in a total volume of 25 μl for 30 min at room temperature. Free DNA was then degraded with 1 U of DNase I (Promega) for exactly 1 min. After addition of 25 μl of 2× stop solution (200 mM NaCl, 200 mM KCl, 20 mM EDTA, 1% sodium dodecyl sulfate, 100 μg of tRNA per ml), the DNA was extracted with phenol-chloroform and chloroform (once each) and ethanol precipitated. The DNA fragments were resuspended in 95% formamide–20 mM EDTA–0.05% xylene cyanol–0.05% bromphenol blue and separated on an 8% polyacrylamide sequencing gel, together with a chemically sequenced probe (24) to identify the positions of the DNase I fragments.
Luciferase assay.
Cells (1.2 × 106) were seeded into 25-ml flasks (Greiner) and transfected after 24 h with 3 μg of the promoterless pSVOAL plasmid containing the luciferase gene (5) into which the different LTR constructs were cloned as promoters, using the DOTAP liposomal transfection reagent as specified by the manufacturer (Boehringer Mannheim). Transfected cells were incubated in medium without fetal calf serum for 5 h and then transferred to medium supplemented with 10% fetal calf serum. At 24 h later, the cells were washed twice with phosphate-buffered saline and lysed in 250 μl of luciferase lysis buffer [1 M Tricine, 100 mM (MgCO3)4Mg(OH)2, 100 mM MgSO4, 10 mM ATP, 10 mM coenzyme A, 0.5 M EDTA, 1 M dithiothreitol (DTT), (pH 7.8) (Promega)] for 5 min. The cell debris was pelleted, and the supernatant was stored at −70°C. Background light emission of 20 μl of cell lysate was measured for 3 min in a luminometer (LB9505; Berthold). Then 100 μl of luciferin solution [71 μg of luciferin in 20 mM Tricine, 1.07 mM (MgCO3)4Mg(OH)2, 2.67 mM MgSO4, 0.1 mM EDTA, 33.3 mM DTT, 0.53 mM ATP, and 0.27 mM coenzyme A (Promega)] was added to measure the luciferase activity for another 3 min.
RESULTS
HERV-K enhancer complex: binding site and composition.
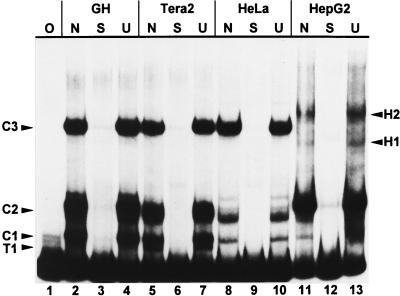
Three regions within the HTDV/HERV-K LTR mainly control expression of the provirus: the 5′ enhancer, the promoter, and the R region. Among these, we started to analyze the regulatory potential of the 5′ enhancer, which roughly comprises the first 170 bp of the U3 region (Fig. 1). The regulatory control exerted by the 5′ enhancer in different cell types should be reflected in a different subset of proteins which bind to the 5′ enhancer depending on whether the cell expresses HERV-K. To test this, EMSAs were performed, with the HERV-K10 (27) LTR 1 to 167 restriction fragment (Fig. 1) as a probe and nuclear protein extracts of the HERV-K-expressing teratocarcinoma cell lines GH and Tera2 as well as of the weakly expressing cervicalcarcinoma HeLa and the nonexpressing hepatocarcinoma HepG2 cell lines (Fig. 2; HERV-K-expressing and weakly or nonexpressing cell lines will be referred to as two different cell types throughout this paper). In addition to three intense common bands (C1, C2, and C3) one high-mobility teratocarcinoma-specific protein complex, T1, and two low-mobility hepatocarcinoma specific complexes, H1 and H2, were observed (Fig. 2). Tera2 extracts always showed a strong T1 band, whereas GH extracts formed only a very weak one. In some HepG2 nuclear extract preparations, the H1 complex resolves into two bands. The HeLa extract, on the other hand, resulted only in bands with identical mobility but different intensities from bands observed with the other cell extracts. The cell-type-specific band shift pattern observed strongly suggested that the 5′ enhancer of HERV-K actively is involved in regulation of the expression of the provirus.
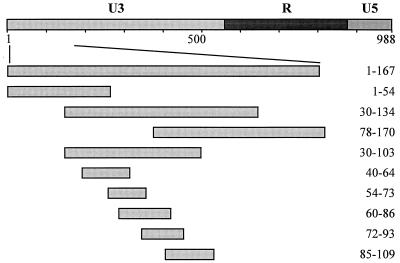
FIG. 1.
Schematic representation of HERV-K LTR and LTR subfragments used for competition EMSAs. U3, R, and U5 regions are represented by different shadings; numbers delineate nucleotide positions in the LTR.
FIG. 2.
EMSA of the HERV-K LTR subfragment from bp 1 to 167 as a probe with nuclear extracts of the cell lines indicated. Lanes: O: no protein extract added; N, no competitor added; S, unlabeled LTR fragment from bp 1 to 167 as specific competitor; U, unlabeled oligonucleotide containing the Sp1 binding site as a nonspecific competitor. All bands are competed specifically. One teratocarcinoma-specific (T1) and two hepatocarcinoma-specific (H1 and H2) complexes are discernible; C1, C2, and C3 are common to all cell lines.
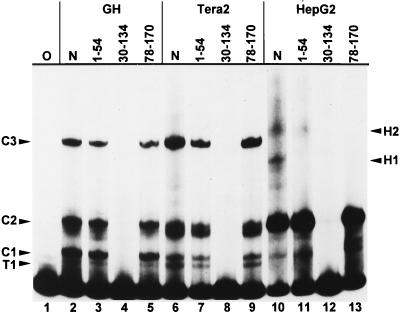
We next were interested in identifying the binding site or sites of the 5′-enhancer protein complexes. For this purpose, overlapping subfragments of the K10 LTR 5′ enhancer were cloned (Fig. 1, 1 to 54, 55 to 167, 30 to 134, 78 to 170, 30 to 103, 78 to 134, 123 to 170) and used as competitors in EMSAs with fragment 1 to 167 as the DNA probe. Competition, i.e., disappearance of bands, indicates that the respective competitor DNA contains the identical protein binding site to the probe. Of these competitors, only subfragments containing HERV-K LTR sequences between bp 55 through 77 (i.e. subfragments 55 to 167, 30 to 134, and 30 to 170) were able to compete all complexes completely (Fig. 3 and data not shown). However, both low-mobility hepatocarcinoma complexes could also be competed with subfragments containing LTR sequences either upstream (H1; 30 to 134) or downstream (H1, H2; 78 to 170) or bp 55 to 77. In addition, when the subfragments 55 to 167, 30 to 134, and 30 to 170 were used as probes, band shift patterns very similar to those obtained with fragment 1 to 167 as the probe emerged, with only minor deviations. Again, all the complexes could be competed only with subfragments containing bp 55 to 77 (data not shown).
FIG. 3.
EMSA of the HERV-K LTR fragment from bp 1 to 167 as a probe with nuclear cell extracts and different LTR subfragments as competitors. Lanes: O, no protein extract added; N, no competitor added. Other lanes contain LTR subfragments as specific competitors; the nucleotide positions are indicated (Fig. 1). Complexes are competed with LTR subfragments containing bp 55 to 77. H1 and H2 complexes are also competed with subfragments containing LTR regions upstream of bp 55 (H1) and downstream of bp 77 (H1 and H2).
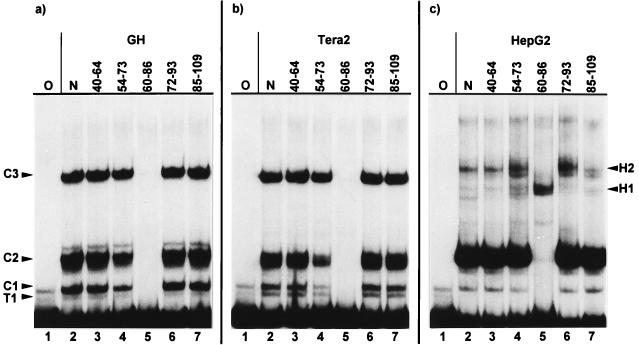
This competition behavior raised the possibility that with the exception of the low-mobility hepatocarcinoma complexes H1 and H2, all other 5′-enhancer complexes assemble at only one or two DNA binding protein(s), which bind within the region between bp 50 and 80. We therefore generated a second set of five smaller HERV-K enhancer subfragments, each about 20 to 25 bp long and overlapping each adjacent one roughly by half (Fig. 1; 40 to 64, 54 to 73, 60 to 86, 72 to 93, and 85 to 109). Using these as competitors in EMSAs with 1 to 167 as a probe, only subfragment 60 to 86 was able to completely compete all complexes of GH, Tera2, and HeLa nuclear extracts (Fig. 4a and b; HeLa not shown). The fact that fragment 54 to 73 already showed a slight competition effect, especially with Tera2 nuclear extracts, indicates that the 3′ part of this fragment harbors sequences relevant but not sufficient for the formation of the complexes. With HepG2 nuclear extracts, however, the H1 complex could not be competed with subfragments 60 to 86; rather, the H1 band became more intense in the presence of this competitor. The other four subfragments competed H1 and H2 only to an extent (Fig. 4c). With the subfragments as a probe, only fragment 60 to 86 generated bands which could be competed specifically, regardless which cell extracts were used (data not shown). GH, Tera2, and HeLa extracts gave the same number of bands with similar relative intensities with both 1 to 167 and 60 to 86 as probes. Both probes also gave the same high-mobility complexes with HepG2 extracts. The HepG2 low-mobility complexes H1 and H2, on the other hand, could not be generated with probe 60 to 86 but could be generated only with longer probes, containing additional LTR sequences adjacent to this 27-bp region (compare Fig. 2, 3, and 4). Moreover, probes 54 to 73, 72 to 93, and 85 to 109 gave specifically competable bands of low mobility only when hepatocarcinoma extracts were used (data not shown), further indicating the importance of LTR sequences abutting the sequence from 60 to 86 for assembly of these HepG2 complexes.
FIG. 4.
EMSA of the HERV-K LTR fragment from bp 1 to 167 as a probe with GH (a), Tera2 (b), and HepG2 (c) nuclear cell extracts and smaller LTR subfragments as competitors. Lanes: O, no protein extract added; N, no competitor added. Other lanes contain specific competitors; the nucleotide positions are indicated (Fig. 1). All complexes of teratocarcinoma cells (a and b) are competed with the LTR subfragment from bp 60 to 86. Most HepG2 complexes are competed by the subfragment containing bp 60 to 86; the intensity of one of the complexes is increased. Some low-mobility complexes of HepG2 cells are competed with LTR sequences adjacent to bp 60 to 86.
To exactly map the binding site of the HERV-K enhancer complex, we applied the DNase I footprinting technique. Both teratocarcinoma and hepatocarcinoma nuclear extracts resulted in a clear footprint between bp 62 and 83 (GGAGACTCCATTTTGTTATGTG) (Fig. 5).
FIG. 5.
DNase I footprint of HERV-K LTR bp 1 to 167. Lanes: 1, no protein extract added; 2 to 4, nuclear extracts of GH, Tera2, and HepG2 cells added, respectively. The vertical bar earmarks the protected region between bp 62 and 83.
In summary, three lines of evidence indicate that the 22-bp element between bp 62 and 83 is the major HERV-K 5′-enhancer complex binding site. First, all protein complexes could be competed with competitor DNA containing contiguous sequences either between bp 55 and 77 or between bp 60 and 86. Second, all teratocarcinoma cell and HeLa cell complexes, as well as the high-mobility HepG2 cell complexes, could be generated on DNA probes containing these sequences. Third, a DNase I footprint was obtained with nuclear extracts between bp 62 and 83. Taken together, these results show that all HERV-K 5′-enhancer complexes bind to and assemble at the site between bp 62 and 83 in both cell types. The EMSA data clearly indicated that in HepG2 cells, although not detectable by DNase I footprinting, sequences adjacent to this binding site, presumably covering the region from bp 50 to 110, contribute important determinants the assembly of the whole enhancer complex in this cell line. Furthermore, the enhancer complexes of HERV-K-expressing and nonexpressing cell types do contain at least partly different protein components.
YY1 is the HERV-K enhancer binding protein.
Having mapped the binding site of the HERV-K enhancer complex, we were interested in identifying the enhancer binding protein(s). The enhancer sequence was checked for putative transcription factor binding sites in the TRANSFAC database with the search routine TFSEARCH (42), and a consensus sequence for the cellular transcription factor YY1 was found between bp 64 and 80 (AGACTCCATTTTGTTAT) with a homology score of 88.9%. In addition, high scoring consensus sequences were found for C/EBP (CAAT enhancer binding protein; 87.7%) between bp 78 and 90 and for SRY (sex region Y protein; 85.9%) between bp 70 and 81. Among these, the C/EBP consensus sequences does not lie completely within the mapped binding site. Since the subfragment from 72 to 93, containing both C/EBP and SRY binding sites, was not effective as a competitor and did not generate specific EMSA bands with any of the cell extracts tested (see above), we assumed that YY1 was the most likely candidate for being a major HERV-K enhancer binding protein.
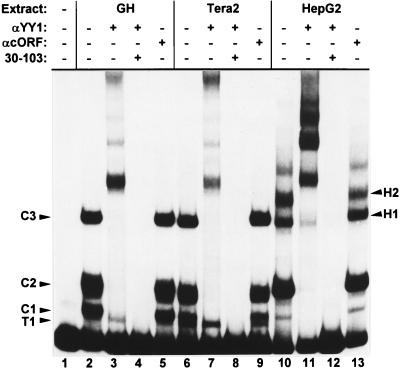
To identify the binding protein, supershift assays were used. In an EMSA with bp 1 to 167 as a probe and nuclear extracts of both cell types, an anti-YY1 antiserum produced at least five supershifted bands with identical mobilities but different intensities depending on the cell type (Fig. 6). With only the binding site from bp 60 to 86 as a probe, the same EMSA bands could be supershifted by anti-YY1 antiserum but resulted in only two supershifted bands (data not shown). The supershifted bands were not caused simply by the addition of serum proteins, as demonstrated by the inclusion in some reaction mixtures of an unrelated antiserum against the HERV-K cORF protein, which did not change the mobility or the intensity of any of the complexes. In addition, no supershift was obtained with an anti-C/EBP antiserum, supporting the exclusion of this transcription factor as a HERV-K enhancer protein (data not shown). Furthermore, all anti-YY1 supershifted bands could be competed specifically with the YY1 binding site containing subfragment 30 to 103. Importantly, the teratocarcinoma-specific T1 complex was the only complex that did not supershift with either probe (Fig. 6). The transcription factor YY1 is present in all cell lines used in this study, as tested by Western blotting (data not shown).
FIG. 6.
Supershift with the HERV-K LTR fragment from bp 1 to 167 protein complexes with an anti-YY1-antibody. Lane 1, no protein extract added; other lanes, anti-YY1-antibody, anti-cORF-antiserum, and subfragment from bp 30 to 103 as a specific competitor were added to individual binding reactions as indicated. Except for complex T1 of both teratocarcinoma cells, all protein complexes are retarded by the anti-YY1-antibody.
Both observations, i.e., the presence of a highly conserved YY1 consensus sequence within the borders of the enhancer binding site and the positive result of the YY1 supershift, strongly suggest that YY1 is the main HERV-K enhancer binding protein. As inferred from the supershift experiments, YY1 is present in all enhancer complexes in HepG2 and HeLa cells, whereas in teratocarcinoma cells an additional and as yet unidentified DNA binding protein, which also binds to the 62 to 83 region of the HERV-K LTR, is present.
YY1 enhances HERV-K expression.
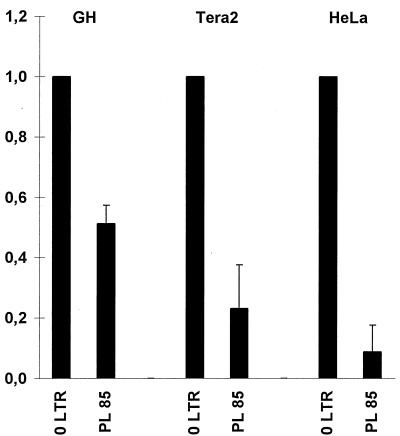
We next wanted to know whether binding of YY1 to the HERV-K enhancer has any effect on the expression of HERV-K. To test this, we completely replaced the YY1 binding site containing the region from bp 61 to 84 with the unrelated sequence from bp 718 to 695 of the pBluescript plasmid polylinker and cloned this otherwise unaltered full-length (988-bp) HERV-K LTR in sense orientation upstream of the luciferase gene as a reporter. This construct, 60PL85, along with the wild-type HERV-K LTR as a control, was transiently transfected in GH, Tera2, HepG2, and HeLa cells. The luciferase activity obtained was determined relative to the activity evoked by the wild-type LTR and taken as a measure of the promoter activity of the mutant LTR. However, since the luciferase counts of both wild-type and mutant LTR measured with HepG2 cell lysates were in the background range of the luminometer (≈104 counts), presumably due to strong repression of the LTR, the HeLa cell line was chosen to represent HERV-K-nonexpressing cells. Figure 7 shows the relative luciferase activity for GH, Tera2, and HeLa cells. The mutant LTR construct resulted in a strong reduction of activity compared to the wild-type LTR activity. The decrease in activity was statistically significant on the basis of _t_-test analysis. With this mutated enhancer as a probe in an EMSA, the nuclear extracts of the cell lines used in the luciferase assay resulted in only a few weak and nonspecific bands; furthermore, no YY1 supershift could be obtained (data not shown). Hence, the loss of specific EMSA bands correlates well with a decrease in luciferase activity. These results demonstrate that YY1 acts as an activator of HERV-K expression in both HERV-K-expressing and nonexpressing cell types.
FIG. 7.
Relative luciferase activity of HERV-K LTR constructs carrying wild-type and mutant YY1 binding sites. Each column represents the mean relative luciferase activity of the LTR constructs indicated. Luciferase counts were measured in triplicate in four independent transfections into Tera2 and HeLa cells. The activity of the wild-type HERV-K LTR was set as 1. The standard deviation is indicated by the error bars. The decrease in luciferase activity is statistically significant according to the t test.
DISCUSSION
HERV-K is expressed in a cell-type-specific manner: whereas most human tissues show only basal expression, detectable by a sensitive reverse transcriptase PCR approach (28a), teratocarcinoma cell lines display high levels of expression in a complex pattern of four distinct mRNA species, as can be seen in Northern blots (19). Luciferase reporter gene assays identified three regions within the 5′ LTR of HERV-K with a profound influence on the promoter activity: the 5′ terminus of the LTR (termed the 5′ enhancer), the promoter region, and the R region (11). Among these, we started to investigate the 5′ enhancer. We identified a YY1 binding site between bp 62 and 83 of the HERV-K LTR and showed that YY1 is the major HERV-K enhancer binding protein in human teratocarcinoma (GH and Tera2), hepatocarcinoma (HepG2), and cervical carcinoma (HeLa) cells. Furthermore, in functional reporter gene assays, we showed that the YY1 enhancer complexes activate HERV-K expression irrespective of the cell type.
The cellular protein YY1 is a 414-amino-acid zinc finger-type transcription factor with four C-terminally located C2-H2-type fingers (32). YY1 is highly conserved from frogs to humans (29) and is ubiquitously expressed in pluripotent as well as differentiated cells, including the mouse teratocarcinoma cell line F9 and HeLa cells (2, 32). The range of YY1-expressing cells has been expanded here to the human teratocarcinoma cell lines GH and Tera2 and the hepatocarcinoma cell line HepG2. It is well established that YY1, together with different cofactors, mediates activation, repression, or initiation of transcription depending on the genes regulated by YY1 (see reference 33 for a compilation). Besides its effect on expression of cellular genes, it regulates viral systems as well. Human immunodeficiency virus type 1, for example, is repressed by YY1 (23). In addition, several retroelements are regulated (activated in this case) by YY1; these include the human long interspersed element LINE-1 (3, 34) and the mouse IAP (31). Regulatory sequences of LINE-1 elements, which do not contain LTRs, are located within the 5′ untranslated region of the transcription unit, i.e., at the 5′ end of the element itself (38). It is from within the first 20 bp of these elements that YY1 activates LINE-1 expression (3). Mouse IAPs do carry LTRs, which contain an IAP upstream enhancer element, located 180 bp upstream of the transcription start site, i.e., within the U3 region. YY1 binds to this enhancer element to stimulate IAP transcription (31). YY1, most notably, is even able to act as both an activator and repressor on the very same gene. The adeno-associated virus P5 promoter normally is repressed by YY1; however, after association with the adenovirus transactivator E1A, it acts as an activator on the P5 promoter (15).
For HTDV/HERV-K, the EMSA pattern obtained with nuclear extracts of HERV-K-expressing GH and Tera2 cells, as opposed to poorly expressing HeLa cells and nonexpressing HepG2 cells, showed that these cell types do have a partly different set of protein factors constituting the enhancer complexes. Teratocarcinoma cells strongly express HERV-K and form an additional, teratocarcinoma-specific complex in EMSAs (T1) (Fig. 1). However, expression is more elevated in GH cells than in Tera2 cells, but the T1 complex is more intense with Tera2 extracts. Two specific complexes (H1 and H2) are seen with HepG2 nuclear extracts, the cell line not expressing HERV-K at all. HeLa cells display an intermediate level of HERV-K expression and have only enhancer complexes common to all cell lines tested (C1, C2, and C3). It thus appears that the presence or absence of certain protein complexes, i.e., the protein composition of the 5′ enhancer complexes, roughly correlates with the magnitude of HERV-K expression in each cell line. These different complexes might exert functionally different effects upon the HERV-K LTR promoter activity. However, to verify this hypothesis, more cell lines must be examined. The enhancer complexes in GH, Tera2, and HeLa cells all seem to bind only between bp 62 and 83, since there is a complete competition of these complexes with the subfragment from bp 60 to 86 (Fig. 4). Furthermore, when used as a probe, this subfragment generates the same bands as the fragment containing the complete enhancer (bp 1 to 167 [data not shown]). The behavior of both HepG2-specific complexes (H1 and H2) is more complex, and neither can be generated in EMSAs with probe bp 60 to 86. This indicates that sequences adjacent to the YY1 binding site may contain important determinants for assembly of the complete enhancer complex in HepG2 cells. However, besides YY1, no further components of the enhancer complexes have been identified. Furthermore, preliminary experiments indicated that the C3 complex consists of the full-length YY1 protein bound to the enhancer. In this case, the C1 and C2 complexes are likely to consist of YY1 degradation products. The H2 complex may contain YY1 and a second protein component, and the H1 complex may consist of a YY1 degradation product and a second protein. Retained DNA binding capacity has been observed with YY1 degradation products in cellular protein extracts and with YY1 proteins containing artificially introduced deletions (2, 25).
Recently, the region from bp 60 to 87 of the HERV-K enhancer has been mapped by DNase I protection with Jurkat nuclear extracts (1). This region is slightly larger than the footprint we obtained. In UV cross-linking experiments, three proteins of 81, 91, and 98 kDa, tentatively designated ERF1, ERF2, and ERF3, respectively, have been found to constitute the most intense band found in EMSAs with Jurkat extracts. However, none of these proteins has been identified, and all are significantly larger than YY1 (68 kDa). Furthermore, the effect of these ERFs on HERV-K expression is also not known. It thus remains to be determined whether expression of HERV-K is subject to a different mode of regulation in T-cell lines from that in teratocarcinoma cell lines.
The YY1 binding site between bp 62 and 83 overlaps with highly conserved consensus binding sites for C/EBP and SRY. Binding of these transcription factors to the HERV-K enhancer, as discussed above, is rather unlikely. Furthermore, a sequence which also overlaps the YY1 binding site has been described as a putative glucocorticoid responsive element (GRE) located between bp 75 and 80 (27). As with many other retroviruses, HERV-K expression can be stimulated by steroid hormones, suggesting steroid receptor binding to the HERV-K LTR (28). However, bp 75 to 80 contains only half of a GRE and thus is most probably not recognized by glucocorticoid receptors (8). Accordingly, no EMSA band could be supershifted with an anti-GR immunoglobulin G antibody (Santa Cruz) in preliminary experiments. Furthermore, this half-site element was not identified in our TRANSFAC database search, whereas three other complete GREs were found at bp 18 to 33, bp 194 to 209, and bp 388 to 403, all of which reside within the U3 region. We therefore suggest that one or more of these GREs are responsible for the steroid hormone stimulation of expression.
Since the identity of the proteins of the HERV-K enhancer complexes, except for YY1 itself, is still elusive, the relative contributions of each single component to the functional enhancer complexes could not be determined. Luciferase assays, performed with the mutant YY1 binding-site construct (Fig. 7), suggested that the YY1 enhancer complexes act as activators irrespective of the cell type. The competition behavior of some EMSA bands obtained with nuclear extracts of HepG2 cells indicated that sequences outside bp 62 to 83 provide determinants for complete assembly of the protein complex in HepG2 cells. However, in the constructs tested in the luciferase assay, only the YY1 binding site itself has been mutated. We therefore cannot rule out the possibility that some HepG2 enhancer complexes are repressive.
On the YY1 binding-site construct, 60PL85, neither the YY1 complexes nor the T1 complex assembled, as tested by EMSA (data not shown). Since the luciferase activity was reduced by 60PL85 to approximately the same extent in GH, Tera2, and HeLa cell lines (Fig. 7) and T1 is not present in HeLa cells, YY1 alone appears to be the main stimulating protein of the HERV-K enhancer. In all four cell lines tested, the 60PL85 construct never resulted in an elevated level of luciferase activity above the reference activity of the wild-type LTR. This suggests that YY1 is an activator even in cell lines with only basal HERV-K expression. However, the luciferase assays also support the notion that the YY1 enhancer complex alone cannot be responsible for the cell-type-specific nature of HERV-K expression. The difference in LTR activity between HERV-K-expressing and nonexpressing cell types is not evident from Fig. 7, which gives only the relative luciferase activities of the mutant constructs relative to the activity of the wild-type LTR in the same cell line. However, the absolute luciferase counts measured clearly reflect the cell-type-specific influence integrated over the full-length LTR: in teratocarcinoma cells, the wild-type HERV-K LTR evoked a luciferase activity on the order of ≈107 to 108 counts as opposed to ≈106 counts in HeLa cells and only ≈104 counts in HepG2 cells (which is within the background range of the luminometer). Using a luciferase construct with a Rous sarcoma virus LTR promoter as the internal control, we could show that these differences are not due to differences in transfection efficiencies.
In addition, we have cloned many full-length HERV-K LTRs differing in sequence from the human genome and tested them for promoter activity by the luciferase assay (unpublished data). Some of them were active as promoters, whereas others resulted in no luciferase activity even in teratocarcinoma cell lines (unpublished data). Within the YY1 binding site, as determined by DNase I footprinting, the sequences of all LTRs differed in only one or three positions with respect to the HERV-K10 LTR sequence. The YY1 core consensus sequence (CCATNTT) (33), however, is identical in all HERV-K LTRs. Moreover, the EMSA pattern, the competition behavior, and the YY1 supershift pattern of different active and inactive LTRs were identical to the ones observed with the K10 enhancer sequence (data not shown [Fig. 2, 3, and 6 give results for K10]). This also indicates that the YY1 enhancer complex is not involved in transcriptional repression of HERV-K. We therefore believe that repression is mediated by other regions of the LTR, e.g., the promoter region, which outweigh the activating effect of the enhancer region in cell lines which do not express HERV-K.
In this report, we have shown that the human endogenous retrovirus HERV-K is activated by an YY1-enhancer complex which assembles at the very 5′ end of the U3 region. To gain further insight into the complex regulation of HERV-K expression, it will be necessary to identify the additional components of the 5′-enhancer complexes present in expressing versus nonexpressing cell lines and to functionally characterize their contribution to the enhancing effect of the complexes.
ACKNOWLEDGMENTS
We are indebted to A. Hornung, H. Bartel, and H. Rahmouni for excellent technical assistance. We thank B. Kaiser, C. Magin, M. Marschall, R. Kurth, R. Tönjes, and J. Denner for stimulating discussions.
REFERENCES
- 1.Akopov S B, Nikolaev L G, Khil P P, Lebedev Y B, Sverdlov E D. Long terminal repeats of human endogenous retrovirus K family (HERV-K) specifically bind host cell nuclear proteins. FEBS Lett. 1998;421:229–233. doi: 10.1016/s0014-5793(97)01569-x. [DOI] [PubMed] [Google Scholar]
- 2.Austen M, Lüscher B, Lüscher-Firzlaff J M. Characterization of the transcriptional regulator YY1. J Biol Chem. 1997;272:1709–1717. doi: 10.1074/jbc.272.3.1709. [DOI] [PubMed] [Google Scholar]
- 3.Becker K G, Swergold G D, Ozato K, Thayer R E. Binding of the ubiquitous nuclear transcription factor YY1 to a cis regulatory sequence in the human LINE-1 transposable element. Hum Mol Genet. 1993;2:1697–1702. doi: 10.1093/hmg/2.10.1697. [DOI] [PubMed] [Google Scholar]
- 4.Boller K, König H, Sauter M, Müller-Lantzsch N, Löwer R, Kurth R. Evidence that HERV-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma-derived retrovirus HTDV. Virology. 1993;196:349–353. doi: 10.1006/viro.1993.1487. [DOI] [PubMed] [Google Scholar]
- 5.DeWet J R, Wood K V, DeLuca M, Helinski D R, Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feuchter-Murthy A E, Freeman J D, Mager D L. Splicing of a human endogenous retrovirus to a novel phospholipase A2 related gene. Nucleic Acids Res. 1993;21:135–143. doi: 10.1093/nar/21.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hard T, Kellenbach E, Boelens R, Maler B A, Dahlman K, Freedman L P, Carlstedt-Duke J, Yamamoto K R, Gustafsson J A, Kaptein R. Solution structure of the glucocorticoid receptor DNA-binding domain. Science. 1990;249:157–160. doi: 10.1126/science.2115209. [DOI] [PubMed] [Google Scholar]
- 9.Kato N, Pfeiffer-Ohlsson S, Kato M, Larsson E, Rydnert J, Ohlsson R, Cohen M. Tissue-specific expression of human provirus ERV3 mRNA in human placenta: two of the three ERV3 mRNAs contain cellular sequences. J Virol. 1987;61:2182–2191. doi: 10.1128/jvi.61.7.2182-2191.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kingsman S M, Kingsman A J. The regulation of human immunodeficiency virus type-1 gene expression. Eur J Biochem. 1996;240:491–507. doi: 10.1111/j.1432-1033.1996.0491h.x. [DOI] [PubMed] [Google Scholar]
- 11.Knössl, M., K. Thelen, R. Löwer, and J. Löwer. Unpublished data.
- 12.Kongsuwan K, Allen J, Adams J M. Expression of Hox-2.4 homeobox gene directed by proviral insertion in a myeloid leukemia. Nucleic Acids Res. 1989;17:1881–1892. doi: 10.1093/nar/17.5.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.La Mantia G, Majello B, Di Cristofano A, Strazzullo M, Minchiotti G, Lania L. Identification of regulatory elements within the minimal promoter region of the human endogenous ERV9 provirus: accurate transcription initiation is controlled by an Inr-like element. Nucleic Acids Res. 1992;20:4129–4139. doi: 10.1093/nar/20.16.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larsson E, Kato N, Cohen M. Human endogenous proviruses. Curr Top Microbiol Immunol. 1989;148:115–132. doi: 10.1007/978-3-642-74700-7_4. [DOI] [PubMed] [Google Scholar]
- 15.Lee J S, Galvin K M, See R H, Eckner R, Livingston D, Moran E, Shi Y. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev. 1995;9:1188–1198. doi: 10.1101/gad.9.10.1188. [DOI] [PubMed] [Google Scholar]
- 16.Leib-Mösch C, Haltmeier M, Werner T, Geigl E M, Brack W R, Francke U, Erfle V, Hehlmann R. Genomic distribution and transcription of solitary HERV-K LTRs. Genomics. 1993;18:261–269. doi: 10.1006/geno.1993.1464. [DOI] [PubMed] [Google Scholar]
- 17.Löwer R, Löwer J, Frank H, Harzmann R, Kurth R. Human teratocarcinomas cultured in vitro produce unique retrovirus-like viruses. J Gen Virol. 1984;65:887–898. doi: 10.1099/0022-1317-65-5-887. [DOI] [PubMed] [Google Scholar]
- 18.Löwer R, Löwer J, Tondera-Koch C, Kurth R. A general method for the identification of transcribed retrovirus sequences (R-U5 PCR) reveals the expression of the human endogenous retrovirus loci HERV-H and HERV-K in teratocarcinoma cells. Virology. 1993;192:501–511. doi: 10.1006/viro.1993.1066. [DOI] [PubMed] [Google Scholar]
- 19.Löwer R, Boller K, Hasenmaier B, Korbmacher C, Müller-Lantzsch N, Löwer J, Kurth R. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc Natl Acad Sci USA. 1993;90:4480–4484. doi: 10.1073/pnas.90.10.4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Löwer R, Tönjes R R, Korbmacher C, Kurth R, Löwer J. Identification of a Rev-related protein by analysis of spliced transcripts of the human endogenous retroviruses HTDV/HERV-K. J Virol. 1995;69:141–149. doi: 10.1128/jvi.69.1.141-149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Löwer R, Löwer J, Kurth R. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci USA. 1996;93:5177–5184. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luciw P A, Leung N J. Mechanisms of retrovirus replication. In: Levy J A, editor. The Retroviridae. Vol. 1. New York, N.Y: Plenum Press; 1992. pp. 159–298. [Google Scholar]
- 23.Margolis D M, Somasundaran M, Green M R. Human transcription factor YY1 represses human immunodeficiency virus type 1 transcription and virion production. J Virol. 1994;68:905–910. doi: 10.1128/jvi.68.2.905-910.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maxam A M, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 25.Mills L K, Shi Y, Millette R L. YY1 is the cellular factor shown previously to bind to regulatory regions of several leaky-late (bc, c1) genes of herpes simplex virus type 1. J Virol. 1994;68:1234–1238. doi: 10.1128/jvi.68.2.1234-1238.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ono M. Molecular Cloning and long terminal repeat sequences of human endogenous retrovirus genes related to types A and B retrovirus genes. J Virol. 1986;58:937–944. doi: 10.1128/jvi.58.3.937-944.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ono M, Yasunaga T, Miyata T, Ushikubo H. Nucleotide sequence of human endogenous retrovirus genome related to the mouse mammary tumor virus genome. J Virol. 1986;60:589–598. doi: 10.1128/jvi.60.2.589-598.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ono M, Kawakami M, Ushikubo H. Stimulation of expression of the human endogenous retrovirus genome by female steroid hormones in human breast cancer cell line T47D. J Virol. 1987;61:2059–2062. doi: 10.1128/jvi.61.6.2059-2062.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Phelps et al. Submitted for publication.
- 29.Pisaneschi G, Ceccotti S, Falchetti M L, Fiumicino S, Carnevali F, Beccari E. Characterization of FIII/YY1, a Xenopus laevis conserved zinc-finger protein binding to the first exon of L1 and L14 ribosomal protein genes. Biochem Biophys Res Commun. 1994;205:1236–1242. doi: 10.1006/bbrc.1994.2797. [DOI] [PubMed] [Google Scholar]
- 30.Ruddell A. Transcription regulatory elements of the avian retroviral long terminal repeat. Virology. 1995;206:1–7. doi: 10.1016/s0042-6822(95)80013-1. [DOI] [PubMed] [Google Scholar]
- 31.Satyamoorthy K, Park K, Atchison M L, Howe C C. The intracisternal A-particle upstream element interacts with transcription factor YY1 to activate transcription: pleiotropic effects of YY1 on distinct DNA promoter elements. Mol Cell Biol. 1993;13:6621–6628. doi: 10.1128/mcb.13.11.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi Y, Seto E, Chang L S, Shenk T. Transcriptional repression by YY1, a human GLI-Krüppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 33.Shrivastava A, Calame K. An analysis of genes regulated by the multi-functional transcriptional regulator Yin Yang-1. Nucleic Acids Res. 1994;22:5151–5155. doi: 10.1093/nar/22.24.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singer M F, Krek V, McMillan J P, Swergold G D, Thayer R E. LINE-1: a human transposable element. Gene. 1993;135:183–188. doi: 10.1016/0378-1119(93)90064-a. [DOI] [PubMed] [Google Scholar]
- 35.Sjøttem E, Anderssen S, Johansen T. The promoter activity of long terminal repeats of the HERV-H family of human retrovirus-like elements is critically dependent on Sp1 family proteins interacting with a GC/GT box located immediately 3′ to the TATA box. J Virol. 1996;70:188–198. doi: 10.1128/jvi.70.1.188-198.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steinhuber S, Brack M, Hunsmann G, Schwelberger H, Dierich M P, Vogetseder W. Distribution of human endogenous retrovirus HERV-K genomes in humans and different primates. Hum Genet. 1995;96:188–192. [Google Scholar]
- 37.Strazzullo M, Majello B, Lania L, La Mantia G. Mutational analysis of the human endogenous ERV9 proviruses promoter region. Virology. 1994;200:686–695. doi: 10.1006/viro.1994.1232. [DOI] [PubMed] [Google Scholar]
- 38.Swergold G D. Identification, characterization and cell specificity of a human LINE-1 promoter. Mol Cell Biol. 1990;10:6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ting C-N, Rosenberg M P, Snow C M, Samuelson L C, Meisler M H. Endogenous retroviral sequences are required for tissue-specific expression of a human salivary amylase gene. Genes Dev. 1992;6:1457–1465. doi: 10.1101/gad.6.8.1457. [DOI] [PubMed] [Google Scholar]
- 40.Tönjes, R. R., R. Löwer, K. Boller, J. Denner, B. Hasenmaier, H. Kirsch, H. König, C. Korbmacher, C. Limbach, R. Lugert, R. C. Phelps, J. Scherer, K. Thelen, J. Löwer, and R. Kurth. 1996. HERV-K: The biologically most active human endogenous retrovirus family. J. Acquired Immune Defic. Syndr. Hum. Retrovirol. 13(Suppl. 1):261–267. [DOI] [PubMed]
- 41.Wilkinson D A, Mager D L, Leong J C. Endogenous human retroviruses. In: Levy J A, editor. The Retroviridae. Vol. 3. New York, N.Y: Plenum Press; 1994. pp. 465–535. [Google Scholar]
- 42.Wingender E. Recognition of regulatory regions in genomic sequences. J Biotechnol. 1994;35:273–280. doi: 10.1016/0168-1656(94)90041-8. [DOI] [PubMed] [Google Scholar]